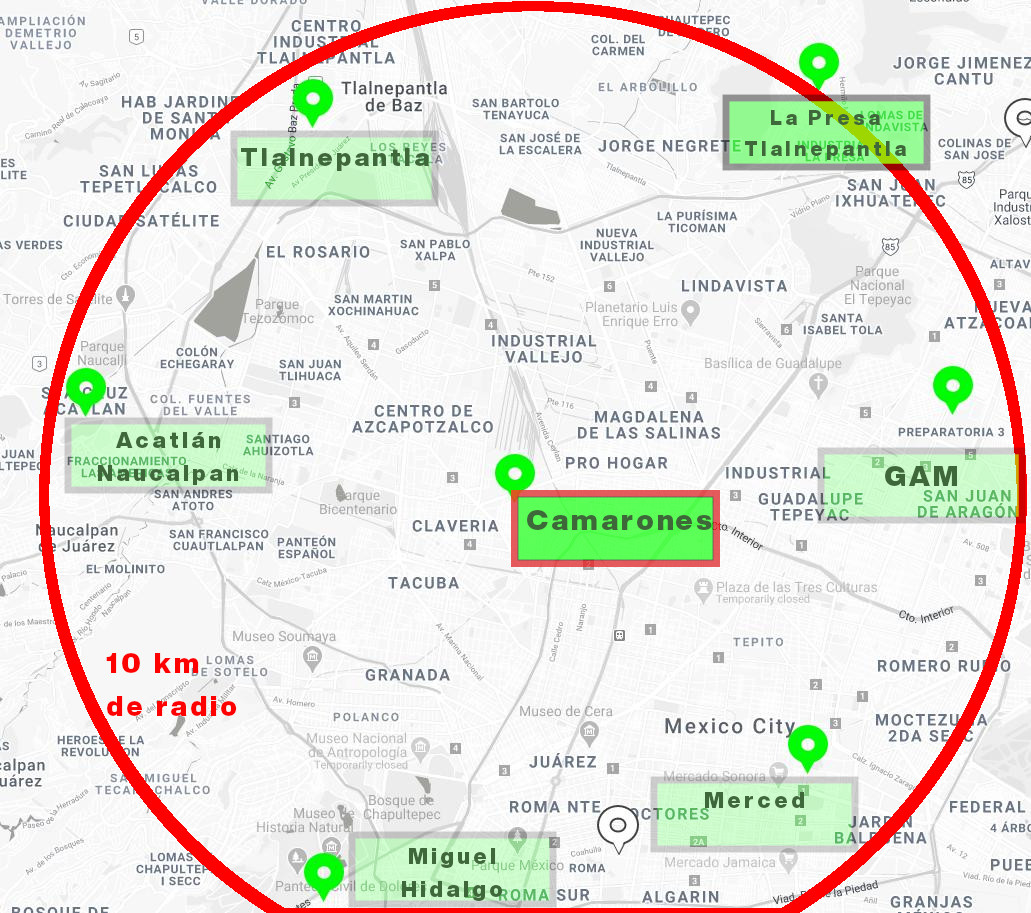
Visualización de las Estaciones Cercanas¶
Encontramos que las siguientes son algunas de las estaciones cercanas a "Camarones" que está próxima a nuestro sensor:
import os, gzip, json, re, stan, dplython, asyncio, nest_asyncio
#nest_asyncio.apply()
import warnings
from matplotlib import pyplot as plt
warnings.filterwarnings("ignore", category=DeprecationWarning)
from dplython import (DplyFrame, X, diamonds, select, sift,
sample_n, sample_frac, head, arrange, mutate, group_by,
summarize, DelayFunction, dfilter)
import seaborn as sns
from plotnine import *
from sklearn.linear_model import LinearRegression, Lasso
from sklearn.model_selection import train_test_split
from sklearn.metrics import (mean_squared_error,
r2_score,
mean_absolute_error)
import pandas as pd
import numpy as np
from IPython.display import display, Markdown
/home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject /home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject /home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject /home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
base_dir = "data/sinaica/"
data_dir = os.path.join(
os.getcwd(),
base_dir
)
#df = pd.read_pickle("data/airdata/air.pickle")
display(Markdown(f"Listing data files from: {data_dir}..."))
generator = (file for file in os.listdir(data_dir)
if (re.match(r"Datos SINAICA - [-A-ZáéíóúÁÉÍÓÚa-z0-9. ]{30,80}\.csv", file) is not None))
sinaica = pd.read_pickle("data/sinaica.pickle")
for (j, file_csv) in enumerate(generator):
#display(Markdown(f"{i+1}. File \"{file_csv}\""))
try:
station, sensor = re.match(
r"Datos SINAICA - ([A-ZáéíóúÁÉÍÓÚa-z. ]{3,20}) - ([A-Z0-9a-z.]+) - [-0-9 ]+\.csv",
file_csv).groups()
#display(Markdown(f" * {station}, {sensor}"))
df2 = pd.read_csv(os.path.join(data_dir, file_csv))
df2 = df2.assign(Estacion=station)
sinaica = pd.concat([sinaica, df2])
#display(df2.head(3))
sinaica = sinaica.sort_values(by=["Fecha", "Estacion", "Parámetro"])
sinaica["Hora"] = sinaica[["Hora"]].replace("- .*$", "", regex=True)
sinaica["Fecha"] = pd.to_datetime(sinaica["Fecha"] + " " + sinaica["Hora"])
#sinaica.drop("Hora", axis=1, inplace=True)
sinaica = sinaica[(sinaica["Fecha"] >= "2021-01-01")].copy()
display(Markdown(f"Done reading {j+1} files!"))
display(pd.concat([sinaica.head(5), sinaica.tail(5)]))
except Exception as e:
print(f"Error while reading file {file_csv}", type(e))
raise e
Listing data files from: /home/jaa6766/Documents/jorge3a/itam/deeplearning/dlfinal/data/sinaica/...
Encontramos que las siguientes son algunas de las estaciones cercanas a "Camarones" que está próxima a nuestro sensor:

(
ggplot(sinaica) +
geom_point(aes(x="Fecha", y="Valor", color="Parámetro")) +
facet_wrap("Parámetro", scales="free") +
labs(title="Visualización general de las Variables de Contaminantes") +
theme(axis_text_x=element_text(angle=90),
subplots_adjust={'wspace': 0.25, 'hspace': 0.25}
)
)
<ggplot: (8794812536053)>
(
ggplot(sinaica[(sinaica["Estacion"] == "Camarones")]) +
geom_point(aes(x="Fecha", y="Valor", color="Parámetro")) +
facet_wrap("Parámetro", scales="free") +
labs(title="Estación Camarones") +
theme(axis_text_x=element_text(angle=90),
subplots_adjust={'wspace': 0.25, 'hspace': 0.25}
)
)
<ggplot: (8794810293949)>
sinaica.to_pickle("data/sinaica/sinaica.pickle")
Algunos datos faltantes son causados por mantenimiento del equipo. Pero pudiéramos enriquecer la información con los datos de estaciones cercanas. Para ello vamos a evaluar las imputaciones.
dsinaica = DplyFrame(pd.read_pickle("data/sinaica/dsinaica.pickle"))
dsinaica
| Fecha | Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | FES Acatlán_CO | ... | Miguel Hidalgo_O3 | Miguel Hidalgo_SO2 | Tlalnepantla_CO | Tlalnepantla_NO | Tlalnepantla_NO2 | Tlalnepantla_NOx | Tlalnepantla_O3 | Tlalnepantla_PM10 | Tlalnepantla_PM2.5 | Tlalnepantla_SO2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2021-01-01 00:00:00 | 0.6 | 0.006 | 0.029 | 0.034 | 0.011 | NaN | NaN | 0.002 | 0.4 | ... | 0.009 | 0.003 | 0.6 | NaN | 0.030 | 0.034 | 0.012 | 37.0 | 19.0 | 0.002 |
| 1 | 2021-01-01 01:00:00 | 1.0 | 0.021 | 0.038 | 0.059 | 0.002 | NaN | NaN | 0.002 | 0.6 | ... | 0.006 | 0.003 | 0.6 | NaN | 0.026 | 0.029 | 0.013 | 42.0 | 29.0 | 0.003 |
| 2 | 2021-01-01 02:00:00 | 0.8 | 0.013 | 0.035 | 0.049 | 0.003 | NaN | NaN | 0.001 | 0.9 | ... | 0.003 | 0.002 | 0.7 | NaN | 0.032 | 0.036 | 0.006 | 58.0 | 43.0 | 0.002 |
| 3 | 2021-01-01 03:00:00 | 1.0 | 0.031 | 0.034 | 0.065 | 0.002 | NaN | NaN | 0.001 | 0.8 | ... | 0.004 | 0.002 | 0.7 | NaN | 0.033 | 0.039 | 0.004 | 59.0 | 41.0 | 0.002 |
| 4 | 2021-01-01 04:00:00 | 0.6 | 0.005 | 0.029 | 0.034 | 0.005 | NaN | NaN | 0.001 | 1.0 | ... | 0.006 | 0.002 | 0.7 | NaN | 0.032 | 0.038 | 0.004 | 64.0 | 46.0 | 0.002 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2166 | 2021-04-01 19:00:00 | 0.4 | 0.003 | 0.011 | 0.013 | 0.016 | 69.0 | 7.0 | 0.001 | 0.3 | ... | 0.023 | 0.000 | 0.3 | 0.009 | 0.017 | 0.025 | 0.017 | 52.0 | 24.0 | 0.003 |
| 2167 | 2021-04-01 20:00:00 | 0.4 | 0.002 | 0.011 | 0.012 | 0.018 | 71.0 | 9.0 | 0.001 | 0.2 | ... | 0.024 | 0.000 | 0.4 | 0.004 | 0.015 | 0.019 | 0.020 | 22.0 | 10.0 | 0.005 |
| 2168 | 2021-04-01 21:00:00 | 0.4 | 0.002 | 0.013 | 0.015 | 0.016 | 37.0 | 9.0 | 0.001 | 0.2 | ... | 0.023 | 0.000 | 0.3 | 0.002 | 0.014 | 0.017 | 0.021 | 21.0 | 14.0 | 0.002 |
| 2169 | 2021-04-01 22:00:00 | 0.4 | 0.002 | 0.019 | 0.021 | 0.012 | 19.0 | 0.0 | 0.001 | 0.1 | ... | 0.027 | 0.000 | 0.3 | 0.002 | 0.011 | 0.013 | 0.022 | 11.0 | 7.0 | 0.001 |
| 2170 | 2021-04-01 23:00:00 | 0.4 | 0.001 | 0.014 | 0.015 | 0.021 | 61.0 | 21.0 | 0.001 | 0.2 | ... | 0.033 | 0.001 | 0.3 | 0.001 | 0.014 | 0.014 | 0.029 | 55.0 | 22.0 | 0.002 |
2171 rows × 45 columns
dsinaica[dsinaica.isna().any(axis=1)]
| Fecha | Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | FES Acatlán_CO | ... | Miguel Hidalgo_O3 | Miguel Hidalgo_SO2 | Tlalnepantla_CO | Tlalnepantla_NO | Tlalnepantla_NO2 | Tlalnepantla_NOx | Tlalnepantla_O3 | Tlalnepantla_PM10 | Tlalnepantla_PM2.5 | Tlalnepantla_SO2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2021-01-01 00:00:00 | 0.6 | 0.006 | 0.029 | 0.034 | 0.011 | NaN | NaN | 0.002 | 0.4 | ... | 0.009 | 0.003 | 0.6 | NaN | 0.030 | 0.034 | 0.012 | 37.0 | 19.0 | 0.002 |
| 1 | 2021-01-01 01:00:00 | 1.0 | 0.021 | 0.038 | 0.059 | 0.002 | NaN | NaN | 0.002 | 0.6 | ... | 0.006 | 0.003 | 0.6 | NaN | 0.026 | 0.029 | 0.013 | 42.0 | 29.0 | 0.003 |
| 2 | 2021-01-01 02:00:00 | 0.8 | 0.013 | 0.035 | 0.049 | 0.003 | NaN | NaN | 0.001 | 0.9 | ... | 0.003 | 0.002 | 0.7 | NaN | 0.032 | 0.036 | 0.006 | 58.0 | 43.0 | 0.002 |
| 3 | 2021-01-01 03:00:00 | 1.0 | 0.031 | 0.034 | 0.065 | 0.002 | NaN | NaN | 0.001 | 0.8 | ... | 0.004 | 0.002 | 0.7 | NaN | 0.033 | 0.039 | 0.004 | 59.0 | 41.0 | 0.002 |
| 4 | 2021-01-01 04:00:00 | 0.6 | 0.005 | 0.029 | 0.034 | 0.005 | NaN | NaN | 0.001 | 1.0 | ... | 0.006 | 0.002 | 0.7 | NaN | 0.032 | 0.038 | 0.004 | 64.0 | 46.0 | 0.002 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2136 | 2021-03-31 13:00:00 | 0.4 | 0.004 | 0.017 | 0.020 | 0.053 | 96.0 | 23.0 | 0.002 | NaN | ... | 0.075 | 0.001 | 0.5 | 0.010 | 0.018 | 0.028 | 0.062 | 64.0 | 37.0 | 0.002 |
| 2144 | 2021-03-31 21:00:00 | 0.4 | 0.002 | 0.019 | 0.021 | 0.021 | 42.0 | 13.0 | 0.001 | 0.3 | ... | NaN | NaN | 0.4 | 0.004 | 0.018 | 0.022 | 0.025 | 112.0 | 43.0 | 0.003 |
| 2146 | 2021-03-31 23:00:00 | 0.8 | 0.013 | 0.063 | 0.076 | 0.007 | 69.0 | 26.0 | 0.002 | 0.6 | ... | 0.043 | 0.001 | 0.6 | 0.002 | 0.042 | 0.044 | 0.023 | 41.0 | 24.0 | 0.002 |
| 2164 | 2021-04-01 17:00:00 | 0.4 | 0.003 | 0.009 | 0.012 | 0.022 | 42.0 | 14.0 | 0.001 | 0.2 | ... | 0.031 | 0.000 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2168 | 2021-04-01 21:00:00 | 0.4 | 0.002 | 0.013 | 0.015 | 0.016 | 37.0 | 9.0 | 0.001 | 0.2 | ... | 0.023 | 0.000 | 0.3 | 0.002 | 0.014 | 0.017 | 0.021 | 21.0 | 14.0 | 0.002 |
1513 rows × 45 columns
Observamos que en la estación Camarones, que es la más cercana, todos los indicadores tienen valores faltantes.
camarones = dsinaica[dsinaica.filter(like="Camarones").isna().any(axis=1)].filter(like="Camarones")
camarones
| Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.6 | 0.006 | 0.029 | 0.034 | 0.011 | NaN | NaN | 0.002 |
| 1 | 1.0 | 0.021 | 0.038 | 0.059 | 0.002 | NaN | NaN | 0.002 |
| 2 | 0.8 | 0.013 | 0.035 | 0.049 | 0.003 | NaN | NaN | 0.001 |
| 3 | 1.0 | 0.031 | 0.034 | 0.065 | 0.002 | NaN | NaN | 0.001 |
| 4 | 0.6 | 0.005 | 0.029 | 0.034 | 0.005 | NaN | NaN | 0.001 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2027 | NaN | NaN | NaN | NaN | NaN | 85.0 | 26.0 | NaN |
| 2028 | NaN | NaN | NaN | NaN | NaN | 67.0 | 22.0 | NaN |
| 2029 | NaN | NaN | NaN | NaN | NaN | 63.0 | 28.0 | NaN |
| 2033 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2050 | 0.8 | NaN | NaN | NaN | NaN | 79.0 | 29.0 | NaN |
611 rows × 8 columns
display(Markdown(f"De ahí que tengamos la idea de no perder una porción considerable: " +
f'{100*dsinaica[dsinaica.filter(like="Camarones").isna().any(axis=1)].filter(like="Camarones").shape[0]/dsinaica.shape[0]:.2f}%' +
f' mediante imputación. Vamos a evaluar los varios métodos con el fin de imputar de la manera más acertada.'
))
De ahí que tengamos la idea de no perder una porción considerable: 28.14% mediante imputación. Vamos a evaluar los varios métodos con el fin de imputar de la manera más acertada.
dsinaica[~dsinaica.filter(like="Camarones").isna().any(axis=1)].filter(like="Camarones")
| Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | |
|---|---|---|---|---|---|---|---|---|
| 479 | 0.5 | 0.021 | 0.024 | 0.046 | 0.002 | 30.0 | 14.0 | 0.008 |
| 480 | 0.6 | 0.017 | 0.023 | 0.039 | 0.002 | 28.0 | 16.0 | 0.006 |
| 481 | 0.5 | 0.023 | 0.022 | 0.046 | 0.002 | 38.0 | 26.0 | 0.005 |
| 482 | 0.6 | 0.030 | 0.021 | 0.051 | 0.002 | 35.0 | 21.0 | 0.003 |
| 483 | 0.8 | 0.057 | 0.022 | 0.079 | 0.002 | 26.0 | 12.0 | 0.003 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2166 | 0.4 | 0.003 | 0.011 | 0.013 | 0.016 | 69.0 | 7.0 | 0.001 |
| 2167 | 0.4 | 0.002 | 0.011 | 0.012 | 0.018 | 71.0 | 9.0 | 0.001 |
| 2168 | 0.4 | 0.002 | 0.013 | 0.015 | 0.016 | 37.0 | 9.0 | 0.001 |
| 2169 | 0.4 | 0.002 | 0.019 | 0.021 | 0.012 | 19.0 | 0.0 | 0.001 |
| 2170 | 0.4 | 0.001 | 0.014 | 0.015 | 0.021 | 61.0 | 21.0 | 0.001 |
1560 rows × 8 columns
camarones_full = dsinaica[~dsinaica.filter(like="Camarones").isna().any(axis=1)]
camarones_train, camarones_test = train_test_split(camarones_full,
test_size=0.3, random_state=175904,
shuffle=False
)
camarones_val = dsinaica[dsinaica.filter(like="Camarones").isna().any(axis=1)]
display(Markdown(f"* Observaciones completas: {camarones_full.shape[0]} (100%)."))
display(Markdown(f">* Observaciones completas en el set de entrenamiento: {camarones_train.shape[0]} (~70%)."))
display(Markdown(f">* Observaciones completas en el set de pruebas: {camarones_test.shape[0]} (~30%)."))
display(Markdown(f"* Observaciones incompletas: {camarones_val.shape[0]}."))
- Observaciones completas en el set de entrenamiento: 1092 (~70%).
- Observaciones completas en el set de pruebas: 468 (~30%).
dsinaica[~dsinaica.filter(like="PM10").isna().any(axis=1)].filter(like="PM10")
| Camarones_PM10 | FES Acatlán_PM10 | Gustavo A. Madero_PM10 | Merced_PM10 | Tlalnepantla_PM10 | |
|---|---|---|---|---|---|
| 479 | 30.0 | 15.0 | 33.0 | 34.0 | 32.0 |
| 480 | 28.0 | 11.0 | 32.0 | 29.0 | 24.0 |
| 481 | 38.0 | 15.0 | 28.0 | 33.0 | 41.0 |
| 482 | 35.0 | 15.0 | 25.0 | 32.0 | 20.0 |
| 483 | 26.0 | 15.0 | 31.0 | 27.0 | 19.0 |
| ... | ... | ... | ... | ... | ... |
| 2165 | 71.0 | 178.0 | 69.0 | 49.0 | 90.0 |
| 2166 | 69.0 | 162.0 | 33.0 | 36.0 | 52.0 |
| 2167 | 71.0 | 49.0 | 32.0 | 24.0 | 22.0 |
| 2169 | 19.0 | 14.0 | 19.0 | 21.0 | 11.0 |
| 2170 | 61.0 | 56.0 | 44.0 | 44.0 | 55.0 |
1295 rows × 5 columns
def describe_with_na(dataframe):
dna = dataframe.isna()
dna = dna.astype('int').sum()
dna.name = "NAs"
dataframe = dataframe.describe()
dataframe = dataframe.append(dna)
dataframe = dataframe.T
dataframe.insert(0, "Estacion", dataframe.index)
return dataframe
describe_with_na(camarones_train.filter(regex=r"PM10", axis=1))
| Estacion | count | mean | std | min | 25% | 50% | 75% | max | NAs | |
|---|---|---|---|---|---|---|---|---|---|---|
| Camarones_PM10 | Camarones_PM10 | 1092.0 | 58.029304 | 27.969164 | 0.0 | 41.0 | 55.0 | 71.0 | 437.0 | 0.0 |
| FES Acatlán_PM10 | FES Acatlán_PM10 | 1007.0 | 48.321748 | 32.849493 | 0.0 | 28.0 | 42.0 | 61.0 | 388.0 | 85.0 |
| Gustavo A. Madero_PM10 | Gustavo A. Madero_PM10 | 1029.0 | 52.945578 | 26.732006 | 0.0 | 34.0 | 50.0 | 67.0 | 352.0 | 63.0 |
| Merced_PM10 | Merced_PM10 | 1084.0 | 56.695572 | 21.527185 | 5.0 | 42.0 | 55.0 | 69.0 | 184.0 | 8.0 |
| Tlalnepantla_PM10 | Tlalnepantla_PM10 | 1034.0 | 52.277563 | 29.432439 | 0.0 | 36.0 | 48.0 | 63.0 | 416.0 | 58.0 |
describe_with_na(dsinaica.filter(regex=r"Camarones", axis=1))
| Estacion | count | mean | std | min | 25% | 50% | 75% | max | NAs | |
|---|---|---|---|---|---|---|---|---|---|---|
| Camarones_CO | Camarones_CO | 2050.0 | 0.771805 | 0.430131 | 0.000 | 0.500 | 0.700 | 1.000 | 3.200 | 121.0 |
| Camarones_NO | Camarones_NO | 2042.0 | 0.025181 | 0.045487 | 0.000 | 0.002 | 0.006 | 0.026 | 0.432 | 129.0 |
| Camarones_NO2 | Camarones_NO2 | 2042.0 | 0.031787 | 0.015682 | 0.003 | 0.020 | 0.031 | 0.042 | 0.111 | 129.0 |
| Camarones_NOx | Camarones_NOx | 2042.0 | 0.056961 | 0.054418 | 0.004 | 0.022 | 0.039 | 0.071 | 0.499 | 129.0 |
| Camarones_O3 | Camarones_O3 | 2042.0 | 0.025693 | 0.024107 | 0.001 | 0.004 | 0.018 | 0.042 | 0.103 | 129.0 |
| Camarones_PM10 | Camarones_PM10 | 1616.0 | 58.066213 | 26.906212 | 0.000 | 41.000 | 55.000 | 71.000 | 437.000 | 555.0 |
| Camarones_PM2.5 | Camarones_PM2.5 | 1604.0 | 25.158978 | 12.946419 | 0.000 | 16.000 | 24.000 | 33.000 | 126.000 | 567.0 |
| Camarones_SO2 | Camarones_SO2 | 2042.0 | 0.006598 | 0.013053 | 0.000 | 0.002 | 0.003 | 0.005 | 0.162 | 129.0 |
contaminante = "Camarones_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "FES Acatlán_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "Gustavo A. Madero_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "Tlalnepantla_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "Merced_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
(
ggplot(camarones_train[["Camarones_PM10"]]) +
geom_histogram(aes(x="Camarones_PM10"), bins=100) +
geom_vline(aes(xintercept=camarones_train["Camarones_PM10"].mean()),
color="red") +
labs(title="Histograma de lecturas en Camarones para PM10")
)
<ggplot: (8794811406597)>
(
ggplot(camarones_train[["Gustavo A. Madero_PM10"]]) +
geom_histogram(aes(x="Gustavo A. Madero_PM10"), bins=100) +
geom_vline(aes(xintercept=camarones_train["Gustavo A. Madero_PM10"].mean()),
color="red") +
labs(title="Histograma de lecturas en Gustavo A. Madero para PM10")
)
/home/jaa6766/.conda/envs/cuda/lib/python3.7/site-packages/plotnine/layer.py:372: PlotnineWarning: stat_bin : Removed 63 rows containing non-finite values.
<ggplot: (8794811379665)>
(
ggplot(camarones_train[["Merced_PM10"]]) +
geom_histogram(aes(x="Merced_PM10"), bins=100) +
labs(title="Histograma de lecturas en La Merced para PM10")
)
/home/jaa6766/.conda/envs/cuda/lib/python3.7/site-packages/plotnine/layer.py:372: PlotnineWarning: stat_bin : Removed 8 rows containing non-finite values.
<ggplot: (8794811438021)>
(
ggplot(camarones_train[["FES Acatlán_PM10"]]) +
geom_histogram(aes(x="FES Acatlán_PM10"), bins=100) +
labs(title="Histograma de lecturas en FES Acatlán para PM10")
)
/home/jaa6766/.conda/envs/cuda/lib/python3.7/site-packages/plotnine/layer.py:372: PlotnineWarning: stat_bin : Removed 85 rows containing non-finite values.
<ggplot: (8794812535869)>
(
ggplot(camarones_train[["Tlalnepantla_PM10"]]) +
geom_histogram(aes(x="Tlalnepantla_PM10"), bins=100) +
labs(title="Histograma de lecturas en FES Acatlán para PM10")
)
/home/jaa6766/.conda/envs/cuda/lib/python3.7/site-packages/plotnine/layer.py:372: PlotnineWarning: stat_bin : Removed 58 rows containing non-finite values.
<ggplot: (8794804237865)>
(
ggplot(camarones_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]) +
geom_point(aes(x="Gustavo A. Madero_PM10", y="Camarones_PM10")) +
labs(title="Gráfica PM10 de 2 Estaciones")
)
<ggplot: (8794811389689)>
(
ggplot(camarones_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]) +
geom_point(aes(x="Merced_PM10", y="Camarones_PM10")) +
labs(title="Gráfica PM10 de 2 Estaciones")
)
<ggplot: (8794804137833)>
(
ggplot(camarones_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]) +
geom_point(aes(x="Tlalnepantla_PM10", y="Camarones_PM10")) +
labs(title="Gráfica PM10 de 2 Estaciones")
)
<ggplot: (8794804206453)>
corr = camarones_train.filter(like="PM10", axis=1)[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)].corr()
corr
| Camarones_PM10 | FES Acatlán_PM10 | Gustavo A. Madero_PM10 | Merced_PM10 | Tlalnepantla_PM10 | |
|---|---|---|---|---|---|
| Camarones_PM10 | 1.000000 | 0.585749 | 0.576044 | 0.644909 | 0.636219 |
| FES Acatlán_PM10 | 0.585749 | 1.000000 | 0.561326 | 0.595378 | 0.755438 |
| Gustavo A. Madero_PM10 | 0.576044 | 0.561326 | 1.000000 | 0.781552 | 0.588582 |
| Merced_PM10 | 0.644909 | 0.595378 | 0.781552 | 1.000000 | 0.599567 |
| Tlalnepantla_PM10 | 0.636219 | 0.755438 | 0.588582 | 0.599567 | 1.000000 |
#corr = corr.loc[:, ["PM2.5", "PM10"]]
mask = None
#mask = np.triu(np.ones_like(corr*-1, dtype=bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
#cmap = sns.palettes.blend_palette(["#ffffff", "#000055"],
# n_colors=16,
# as_cmap=True)
cmap = sns.color_palette("light:blue", as_cmap=True)
plt.title("Mapa de Calor de Correlacioness\n" +
"(correlación en valores absolutos).", loc='left')
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr, mask=mask, cmap=cmap, #vmax=.3, center=0,
square=True, linewidths=.5, cbar_kws={"shrink": .5})
#plt.xlabel("Variables Respuesta")
#plt.ylabel("Variables Predictoras")
plt.show()
corr = camarones_train.filter(like="Camarones", axis=1)[~camarones_train.filter(regex=r"Camarones", axis=1).isna().any(axis=1)].corr()
corr
| Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | |
|---|---|---|---|---|---|---|---|---|
| Camarones_CO | 1.000000 | 0.743399 | 0.770512 | 0.840918 | -0.452770 | 0.298775 | 0.424494 | 0.103995 |
| Camarones_NO | 0.743399 | 1.000000 | 0.452126 | 0.968873 | -0.430920 | 0.123757 | 0.202277 | 0.102433 |
| Camarones_NO2 | 0.770512 | 0.452126 | 1.000000 | 0.658736 | -0.557640 | 0.283317 | 0.434776 | 0.160174 |
| Camarones_NOx | 0.840918 | 0.968873 | 0.658736 | 1.000000 | -0.517954 | 0.183213 | 0.291152 | 0.130837 |
| Camarones_O3 | -0.452770 | -0.430920 | -0.557640 | -0.517954 | 1.000000 | 0.030740 | 0.005117 | -0.194188 |
| Camarones_PM10 | 0.298775 | 0.123757 | 0.283317 | 0.183213 | 0.030740 | 1.000000 | 0.612167 | 0.153458 |
| Camarones_PM2.5 | 0.424494 | 0.202277 | 0.434776 | 0.291152 | 0.005117 | 0.612167 | 1.000000 | 0.290375 |
| Camarones_SO2 | 0.103995 | 0.102433 | 0.160174 | 0.130837 | -0.194188 | 0.153458 | 0.290375 | 1.000000 |
#corr = corr.loc[:, ["PM2.5", "PM10"]]
mask = None
#mask = np.triu(np.ones_like(corr*-1, dtype=bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
#cmap = sns.palettes.blend_palette(["#ffffff", "#000055"],
# n_colors=16,
# as_cmap=True)
#cmap = sns.diverging_palette(250, 20, as_cmap=True)
cmap = sns.color_palette("coolwarm", as_cmap=True)
plt.title("Mapa de Calor de Correlacioness\n" +
"(correlación en valores absolutos).", loc='left')
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr, mask=mask, cmap=cmap, #vmax=.3, center=0,
square=True, linewidths=.5, cbar_kws={"shrink": .5})
#plt.xlabel("Variables Respuesta")
#plt.ylabel("Variables Predictoras")
plt.show()
(
ggplot(camarones_train[["Camarones_PM2.5"]]) +
geom_histogram(aes(x="Camarones_PM2.5"), bins=100) +
labs(title="Histograma de lecturas en Camarones para PM2.5")
)
<ggplot: (8794811495697)>
Removemos observaciones incompletas para realizar la regresión.
Y_train = camarones_train["Camarones_PM10"]
Y_test = camarones_test["Camarones_PM10"]
X_train = camarones_train.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
X_test = camarones_test.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
X_train = camarones_train.filter(regex="(Merced|Tlalnepantla)")
X_test = camarones_test.filter(regex="(Merced|Tlalnepantla)")
## guardar los parquet para correr Stan en R
camarones_train.to_parquet("data/camarones_train.parquet")
camarones_test.to_parquet("data/camarones_test.parquet")
### Código para filtrar solo las columnas con PM10
### Es decir, estaciones que tienen el contaminante PM10
X_test = X_test.filter(regex=r"PM10", axis=1)
X_train = X_train.filter(regex=r"PM10", axis=1)
## Removemos observaciones incompletas
X_train = X_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
X_test = X_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_train = Y_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_test = Y_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
reg = LinearRegression(n_jobs=8).fit(X_train, Y_train)
Yhat_train_lin = reg.predict(X_train)
Yhat_test_lin = reg.predict(X_test)
X_train.columns
Index(['Merced_PM10', 'Tlalnepantla_PM10'], dtype='object')
reg.coef_
array([0.53072702, 0.37388491])
reg.intercept_
8.488947860546723
r2_score(Y_train, Yhat_train_lin)
0.5131358450122813
r2_score(Y_test, Yhat_test_lin)
-0.4144452395742104
mean_squared_error(Y_train, Yhat_train_lin)
374.41392923127256
mean_squared_error(Y_test, Yhat_test_lin)
858.5768327522378
mean_absolute_error(Y_train, Yhat_train_lin)
10.999394181631775
mean_absolute_error(Y_test, Yhat_test_lin)
13.10422582517235
(
ggplot(pd.DataFrame({
"Y_train": Y_train,
"Yhat_train": Yhat_train_lin
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8794810327709)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_lin
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8794804295393)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_lin
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8794810329233)>
Y_train = camarones_train["Camarones_PM10"]
Y_test = camarones_test["Camarones_PM10"]
X_train = camarones_train.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
X_test = camarones_test.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
### Código para filtrar solo las columnas con PM10
### Es decir, estaciones que tienen el contaminante PM10
X_test = X_test.filter(regex=r"PM10", axis=1)
X_train = X_train.filter(regex=r"PM10", axis=1)
## Removemos observaciones incompletas
X_train = X_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
X_test = X_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_train = Y_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_test = Y_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
reg = Lasso().fit(X_train, Y_train)
Yhat_train_lasso = reg.predict(X_train)
Yhat_test_lasso = reg.predict(X_test)
X_train.columns
Index(['FES Acatlán_PM10', 'Gustavo A. Madero_PM10', 'Merced_PM10',
'Tlalnepantla_PM10'],
dtype='object')
reg.coef_
array([0.09567696, 0.06840776, 0.4426 , 0.29441044])
reg.intercept_
9.366559256014888
r2_score(Y_train, Yhat_train_lasso)
0.5199968593721296
r2_score(Y_test, Yhat_test_lasso)
-0.1294304409086644
mean_squared_error(Y_train, Yhat_train_lasso)
369.1375922517144
mean_squared_error(Y_test, Yhat_test_lasso)
685.5711226128724
mean_absolute_error(Y_train, Yhat_train_lasso)
10.90154265031963
mean_absolute_error(Y_test, Yhat_test_lasso)
12.558248813984083
(
ggplot(pd.DataFrame({
"Y_train": Y_train,
"Yhat_train": Yhat_train_lasso
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8794811532553)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_lasso
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8794812109353)>
Se utilizó Stan para realizar una aproximación de los datos que se distribuyen similar a una Poisson.
Yhat_train_stan = pd.read_parquet("data/camarones_train_yhat-stan.parquet")
Yhat_train_stan = Yhat_train_stan["mean-all chains"]
Yhat_test_stan = pd.read_parquet("data/camarones_test_yhat-stan.parquet")
Yhat_test_stan = Yhat_test_stan["mean-all chains"]
Yhat_train_stan.shape[0]
882
Y_train.shape[0]
882
Y_test.shape, Yhat_test_stan.shape
((364,), (414,))
r2_score(Y_train, Yhat_train_stan)
0.22337355392189118
mean_squared_error(Y_train, Yhat_train_stan)
597.2502930070067
mean_absolute_error(Y_train, Yhat_train_stan)
12.397323356009071
df = pd.DataFrame({
"Y_test": Y_test.reset_index(drop=True),
"Yhat_test": Yhat_test_stan
})
df = df[~df.isnull().any(axis=1)]
mean_absolute_error(df[["Y_test"]], df[["Yhat_test"]])
49.862637362637365
gdf = pd.concat({
"Y_train": Y_train.reset_index(drop=True),
"Yhat_train": Yhat_train_stan
}, axis=1)[["Y_train", "Yhat_train"]]
gdf
| Y_train | Yhat_train | |
|---|---|---|
| 0 | 30.0 | 42.5550 |
| 1 | 28.0 | 39.7008 |
| 2 | 38.0 | 43.0379 |
| 3 | 35.0 | 40.4369 |
| 4 | 26.0 | 38.4014 |
| ... | ... | ... |
| 877 | 45.0 | 52.5123 |
| 878 | 39.0 | 58.2521 |
| 879 | 69.0 | 70.1079 |
| 880 | 82.0 | 81.5443 |
| 881 | 51.0 | 55.8011 |
882 rows × 2 columns
(
ggplot(pd.DataFrame({
"Y_train": Y_train.reset_index(drop=True),
"Yhat_train": Yhat_train_stan
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"),
color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"),
color='red', alpha=.5)
)
<ggplot: (8794811513161)>
df = pd.DataFrame({
"Y_test": Y_test.reset_index(drop=True),
"Yhat_test": Yhat_test_stan
})
df = df[~df.isnull().any(axis=1)]
(
ggplot(df) +
geom_point(aes(x=range(0, df.shape[0]), y="Y_test"),
color='blue') +
geom_point(aes(x=range(0, df.shape[0]), y="Yhat_test"),
color='red', alpha=.5)
)
<ggplot: (8794811531597)>
Yhat_train_mean = X_train.mean(axis=1)
Yhat_test_mean = X_test.mean(axis=1)
mean_squared_error(Y_train, Yhat_train_mean)
425.91850907029476
mean_squared_error(Y_test, Yhat_test_mean)
581.5554601648352
mean_absolute_error(Y_train, Yhat_train_mean)
12.331632653061224
mean_absolute_error(Y_test, Yhat_test_mean)
13.761675824175825
(
ggplot(pd.DataFrame({
"Y_train": Y_train,
"Yhat_train": Yhat_train_mean
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"),
color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"),
color='red', alpha=.5)
)
<ggplot: (8794811600257)>
(
ggplot(pd.DataFrame({
"Y_train": Y_test,
"Yhat_train": Yhat_test_mean
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"),
color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_train"),
color='red', alpha=.5)
)
<ggplot: (8794811062369)>
from sklearn.linear_model import PoissonRegressor
reg = PoissonRegressor(alpha=1e3)
reg_fit = reg.fit(X_train, Y_train)
Yhat_train_glm = reg_fit.predict(X_train)
(Yhat_train_glm.shape, Y_train.shape)
((882,), (882,))
Yhat_test_glm = reg_fit.predict(X_test)
(Yhat_test_glm.shape, Y_test.shape)
((364,), (364,))
mean_squared_error(Yhat_train_glm
, Y_train)
574.6123333889531
mean_squared_error(Yhat_test_glm, Y_test)
8981.353126604927
mean_absolute_error(Yhat_train_glm, Y_train)
12.397197210039439
mean_absolute_error(Yhat_test_glm, Y_test)
18.54832154876485
(
ggplot(pd.DataFrame({
"Y_train": Y_train.reset_index(drop=True),
"Yhat_train": Yhat_train_glm
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8794810445797)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_glm
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8794811596521)>
from sklearn.neighbors import KNeighborsRegressor
knn = KNeighborsRegressor(n_neighbors=20, p=1)
knn_fit = knn.fit(X_train, Y_train)
Yhat_train_knn = knn_fit.predict(X_train)
(Yhat_train_knn.shape, Y_train.shape)
((882,), (882,))
Yhat_test_knn = knn_fit.predict(X_test)
(Yhat_test_knn.shape, Y_test.shape)
((364,), (364,))
mean_squared_error(Yhat_train_knn,
Y_train)
350.21837018140593
mean_squared_error(Yhat_test_knn, Y_test)
330.019271978022
mean_absolute_error(Yhat_train_knn, Y_train)
10.843934240362811
mean_absolute_error(Yhat_test_knn, Y_test)
11.718681318681318
(
ggplot(pd.DataFrame({
"Y_train": Y_train.reset_index(drop=True),
"Yhat_train": Yhat_train_knn
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8794810910697)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_knn
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8794810901233)>
df = pd.DataFrame({
"Y_test": Y_test.reset_index(drop=True),
"Yhat_test": Yhat_test_stan
})
df = df[~df.isnull().any(axis=1)]
df2 = pd.DataFrame({
"Model": [
"1.2 Regresión Lineal",
"1.3 Lasso",
"1.4 Stan",
"1.5 Media",
"1.6 GLM",
"1.7 KNN"
],
"MSE (Train Set)": [
mean_squared_error(Y_train, Yhat_train_lin),
mean_squared_error(Y_train, Yhat_train_lasso),
mean_squared_error(Y_train, Yhat_train_stan),
mean_squared_error(Y_train, Yhat_train_mean),
mean_squared_error(Y_train, Yhat_train_glm),
mean_squared_error(Y_train, Yhat_train_knn),
],
"MAE (Train Set)": [
mean_absolute_error(Y_train, Yhat_train_lin),
mean_absolute_error(Y_train, Yhat_train_lasso),
mean_absolute_error(Y_train, Yhat_train_stan),
mean_absolute_error(Y_train, Yhat_train_mean),
mean_absolute_error(Y_train, Yhat_train_glm),
mean_absolute_error(Y_train, Yhat_train_knn),
],
"MSE (Test Set)": [
mean_squared_error(Y_test, Yhat_test_lin),
mean_squared_error(Y_test, Yhat_test_lasso),
mean_squared_error(df[["Y_test"]], df[["Yhat_test"]]),
mean_squared_error(Y_test, Yhat_test_mean),
mean_squared_error(Y_test, Yhat_test_glm),
mean_squared_error(Y_test, Yhat_test_knn),
],
"MAE (Test Set)": [
mean_absolute_error(Y_test, Yhat_test_lin),
mean_absolute_error(Y_test, Yhat_test_lasso),
mean_absolute_error(df[["Y_test"]], df[["Yhat_test"]]),
mean_absolute_error(Y_test, Yhat_test_mean),
mean_absolute_error(Y_test, Yhat_test_glm),
mean_absolute_error(Y_test, Yhat_test_knn),
]
})
df2.sort_values(by="MSE (Test Set)")
| Model | MSE (Train Set) | MAE (Train Set) | MSE (Test Set) | MAE (Test Set) | |
|---|---|---|---|---|---|
| 5 | 1.7 KNN | 350.218370 | 10.843934 | 330.019272 | 11.718681 |
| 3 | 1.5 Media | 425.918509 | 12.331633 | 581.555460 | 13.761676 |
| 1 | 1.3 Lasso | 369.137592 | 10.901543 | 685.571123 | 12.558249 |
| 0 | 1.2 Regresión Lineal | 374.413929 | 10.999394 | 858.576833 | 13.104226 |
| 2 | 1.4 Stan | 597.250293 | 12.397323 | 3087.945055 | 49.862637 |
| 4 | 1.6 GLM | 574.612333 | 12.397197 | 8981.353127 | 18.548322 |
Dado que cambiamos para utilizar los datos como secuenciales (serie de tiempo, optamos por utilizar el modelo basado en interpolación lineal.
Por ello, en la siguiente sección lo realizaremos:
Sabemos por la exploración de datos y en secciones anteriores que la Merced tiene datos muy similares a Camarones, además tiene la bondad de tener menos faltantes.
merced = dsinaica.filter(regex='^Merced_').describe()
dsinaica.shape[0] - merced.iloc[0]
Merced_CO 84.0 Merced_NO 790.0 Merced_NO2 101.0 Merced_NOx 101.0 Merced_O3 95.0 Merced_PM10 29.0 Merced_PM2.5 34.0 Merced_SO2 77.0 Name: count, dtype: float64
Por otra parte camarones tiene muchos más:
camarones = dsinaica.filter(regex='^Camarones_').describe()
dsinaica.shape[0] - camarones.iloc[0]
Camarones_CO 121.0 Camarones_NO 129.0 Camarones_NO2 129.0 Camarones_NOx 129.0 Camarones_O3 129.0 Camarones_PM10 555.0 Camarones_PM2.5 567.0 Camarones_SO2 129.0 Name: count, dtype: float64
Por estos resultados haremos las siguientes imputaciones en un nuevo dataframe.
Quedando el dataframe con la columnas de contaminantes de la siguiente manera:
CO: Merced
NO: Camarones
NO2: Merced
NOx: Merced
O3: Merced
PM10: Merced
PM2.5: Merced
Realizamos un lag, para observar las horas faltantes y detectar las interrupciones de la línea de tiempo:
sinaica_imputated = pd.DataFrame(dsinaica["Fecha"])
sinaica_imputated["CO"] = dsinaica["Merced_CO"]
sinaica_imputated["NO"] = dsinaica["Camarones_NO"]
sinaica_imputated["NO2"] = dsinaica["Merced_NO2"]
sinaica_imputated["NOx"] = dsinaica["Merced_NOx"]
sinaica_imputated["O3"] = dsinaica["Merced_O3"]
sinaica_imputated["PM10"] = dsinaica["Merced_PM10"]
sinaica_imputated["PM2.5"] = dsinaica["Merced_PM2.5"]
sinaica_imputated["SO2"] = dsinaica["Merced_SO2"]
sinaica_imputated["datetime"] = pd.to_datetime(sinaica_imputated["Fecha"])
sinaica_imputated = sinaica_imputated[~sinaica_imputated.isna().any(axis=1)]
sinaica_imputated["year"] = [dt.year for dt in sinaica_imputated.datetime]
sinaica_imputated["month"] = [dt.month for dt in sinaica_imputated.datetime]
sinaica_imputated["day"] = [dt.day for dt in sinaica_imputated.datetime]
sinaica_imputated["hour"] = [dt.hour for dt in sinaica_imputated.datetime]
sinaica_imputated["datetime-1"] = sinaica_imputated["datetime"].shift(1)
sinaica_imputated["delta"] = sinaica_imputated["datetime"] - sinaica_imputated["datetime-1"]
sinaica_imputated["delta"] = (sinaica_imputated["delta"].dt.seconds // 60**2)
sinaica_imputated["imputated"] = False
sinaica_imputated = sinaica_imputated[~sinaica_imputated.isna().any(axis=1)]
sinaica_imputated.reset_index(drop=True, inplace=True)
sinaica_imputated
| Fecha | CO | NO | NO2 | NOx | O3 | PM10 | PM2.5 | SO2 | datetime | year | month | day | hour | datetime-1 | delta | imputated | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2021-01-01 02:00:00 | 1.1 | 0.013 | 0.032 | 0.039 | 0.004 | 37.0 | 24.0 | 0.003 | 2021-01-01 02:00:00 | 2021 | 1 | 1 | 2 | 2021-01-01 00:00:00 | 2.0 | False |
| 1 | 2021-01-01 03:00:00 | 1.2 | 0.031 | 0.033 | 0.043 | 0.001 | 49.0 | 39.0 | 0.003 | 2021-01-01 03:00:00 | 2021 | 1 | 1 | 3 | 2021-01-01 02:00:00 | 1.0 | False |
| 2 | 2021-01-01 04:00:00 | 1.2 | 0.005 | 0.031 | 0.039 | 0.002 | 80.0 | 65.0 | 0.003 | 2021-01-01 04:00:00 | 2021 | 1 | 1 | 4 | 2021-01-01 03:00:00 | 1.0 | False |
| 3 | 2021-01-01 05:00:00 | 1.2 | 0.016 | 0.028 | 0.036 | 0.002 | 89.0 | 75.0 | 0.003 | 2021-01-01 05:00:00 | 2021 | 1 | 1 | 5 | 2021-01-01 04:00:00 | 1.0 | False |
| 4 | 2021-01-01 06:00:00 | 1.4 | 0.024 | 0.029 | 0.060 | 0.001 | 75.0 | 64.0 | 0.003 | 2021-01-01 06:00:00 | 2021 | 1 | 1 | 6 | 2021-01-01 05:00:00 | 1.0 | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1945 | 2021-04-01 18:00:00 | 0.7 | 0.003 | 0.012 | 0.021 | 0.020 | 49.0 | 6.0 | 0.001 | 2021-04-01 18:00:00 | 2021 | 4 | 1 | 18 | 2021-04-01 17:00:00 | 1.0 | False |
| 1946 | 2021-04-01 19:00:00 | 0.6 | 0.003 | 0.009 | 0.013 | 0.021 | 36.0 | 6.0 | 0.001 | 2021-04-01 19:00:00 | 2021 | 4 | 1 | 19 | 2021-04-01 18:00:00 | 1.0 | False |
| 1947 | 2021-04-01 20:00:00 | 0.6 | 0.002 | 0.010 | 0.014 | 0.021 | 24.0 | 0.0 | 0.001 | 2021-04-01 20:00:00 | 2021 | 4 | 1 | 20 | 2021-04-01 19:00:00 | 1.0 | False |
| 1948 | 2021-04-01 22:00:00 | 0.6 | 0.002 | 0.012 | 0.016 | 0.019 | 21.0 | 4.0 | 0.001 | 2021-04-01 22:00:00 | 2021 | 4 | 1 | 22 | 2021-04-01 20:00:00 | 2.0 | False |
| 1949 | 2021-04-01 23:00:00 | 0.6 | 0.001 | 0.009 | 0.011 | 0.035 | 44.0 | 17.0 | 0.002 | 2021-04-01 23:00:00 | 2021 | 4 | 1 | 23 | 2021-04-01 22:00:00 | 1.0 | False |
1950 rows × 17 columns
Vemos que las Horas faltantes son como siguen:
(sinaica_imputated[sinaica_imputated.delta > 2].
sort_values("delta", ascending=False)
[["Fecha", "delta"]].
reset_index(drop=True).
rename(columns={"delta": "Horas Faltantes"}).
head(10)
)
| Fecha | Horas Faltantes | |
|---|---|---|
| 0 | 2021-03-17 09:00:00 | 17.0 |
| 1 | 2021-02-26 08:00:00 | 15.0 |
| 2 | 2021-01-09 13:00:00 | 13.0 |
| 3 | 2021-03-11 23:00:00 | 12.0 |
| 4 | 2021-01-21 23:00:00 | 8.0 |
| 5 | 2021-02-22 15:00:00 | 6.0 |
| 6 | 2021-02-26 14:00:00 | 6.0 |
| 7 | 2021-03-22 15:00:00 | 5.0 |
| 8 | 2021-01-01 18:00:00 | 5.0 |
| 9 | 2021-03-31 12:00:00 | 5.0 |
Realizamos una interpolación quedando los datos así:
%%time
def interpolate_missing(df, idx, hours=1):
"""
Function to interpolate missing.
examples
airdata2 = interpolate_missing(airdata, idx = airdata[airdata.delta > 100].index[0])
airdata2 = interpolate_missing(airdata, 194044)
"""
np.random.seed(175904)
df_prev = df.loc[idx - 1]
a = df_prev
b = df.loc[idx]
offsetEnd = pd.offsets.Hour(1) # remove closed form
out = {}
out["datetime"] = pd.date_range(a["datetime"] + offsetEnd,
b["datetime"] - offsetEnd,
freq='1h')
out["datetime"] = out["datetime"].set_names("datetime")
for v in ['CO', 'NO', 'NO2', 'NOx', 'O3', 'PM10', 'PM2.5', 'SO2']:
i = [i+1 for i, d in enumerate(out["datetime"])]
m = (b[v] - a[v])/len(out["datetime"])
sd = 0.7*np.std(df_prev[v])
rnds = np.random.normal(-sd, sd, len(out["datetime"]))
#rnds = np.random.uniform(-2*np.pi, 2*np.pi, len(out["datetime"]))
#rnds = np.cos(rnds) * sd
out[v] = [m*j + a[v] + rnds[j-1] for j in i]
#out[v] = [m*j b[v] + rnds[j-1] for j in i]
#out["iaqAccuracy"] = 1
idf = pd.DataFrame(out)
reorder_columns = [col for col in out.keys() if col != 'datetime']
reorder_columns.append("datetime")
idf = idf.reindex(columns=reorder_columns)
#print(reorder_columns)
idf["year"] = [dt.year for dt in idf["datetime"]]
idf["month"] = [dt.month for dt in idf["datetime"]]
idf["day"] = [dt.day for dt in idf["datetime"]]
idf["hour"] = [dt.hour for dt in idf["datetime"]]
#idf["minute"] = [dt.minute for dt in idf["datetime"]]
idf["imputated"] = True
# original dataframe
idf = pd.concat([df.reset_index(drop=True), idf.reset_index(drop=True)])
idf.sort_values("datetime", inplace=True)
idf.reset_index(inplace=True, drop=True)
idf["datetime-1"] = idf["datetime"].shift(1)
idf["delta"] = idf["datetime"] - idf["datetime-1"]
idf["delta"] = idf["delta"].dt.seconds // 60**2
return idf
sinaica2 = sinaica_imputated.copy()
#sinaica2 = interpolate_missing(sinaica2, 1600)
#sinaica2[sinaica2.datetime >= pd.to_datetime('2021-03-15 09:00:00')][sinaica2.datetime <= pd.to_datetime('2021-03-17 11:00:00')]
imputation_list = [x for x in reversed(sinaica_imputated.delta[sinaica_imputated.delta > 1].index)]
sinaica2 = sinaica_imputated.copy()
for x in imputation_list:
try:
sinaica2 = interpolate_missing(sinaica2, x)
except:
print(f"Skipping {x}")
sinaica2 = sinaica2.loc[1:].reset_index(drop=True)
#sinaica2
Skipping 0 CPU times: user 810 ms, sys: 5.98 ms, total: 816 ms Wall time: 814 ms
airdata = pd.read_pickle("data/airdata/air-imputated.pickle.gz")
#airdata = airdata.groupby(["year", "month", "day", "hour"], as_index=False)
#airdata = airdata.aggregate(np.mean)
airdata = airdata[[c for c in airdata.columns if c not in ["iaqAccuracy", "year"]]]
#airdata["IAQ"] = airdata[["IAQ"]].round(0).astype('int64')
airdata["minute"] = airdata[["IAQ"]].round(0).astype('int64')
airdata = airdata[["datetime", "month", "day", "hour", "minute", "temperature", "pressure", "humidity",
"gasResistance", "IAQ"]].copy()
sinaica3 = sinaica2[[c for c in sinaica2.columns if c not in ["datetime-1", "delta",
"imputated", "year",
"datetime", "Fecha"]]]
sinaica3 = pd.merge(sinaica3, airdata, on=['month', 'day', 'hour'], how="left")
sinaica3 = sinaica3.loc[~sinaica3.isna().any(axis=1)]
sinaica3
| CO | NO | NO2 | NOx | O3 | PM10 | PM2.5 | SO2 | month | day | hour | datetime | minute | temperature | pressure | humidity | gasResistance | IAQ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 987 | 2.2 | 0.205 | 0.031 | 0.207 | 0.002 | 45.0 | 22.0 | 0.004 | 2 | 12 | 6 | 2021-02-12 06:05:35.846304417 | 35.0 | 21.51 | 777.41 | 44.04 | 152149.0 | 34.7 |
| 988 | 2.2 | 0.205 | 0.031 | 0.207 | 0.002 | 45.0 | 22.0 | 0.004 | 2 | 12 | 6 | 2021-02-12 06:05:38.837326527 | 34.0 | 21.51 | 777.41 | 43.98 | 152841.0 | 33.6 |
| 989 | 2.2 | 0.205 | 0.031 | 0.207 | 0.002 | 45.0 | 22.0 | 0.004 | 2 | 12 | 6 | 2021-02-12 06:05:47.812360048 | 32.0 | 21.54 | 777.41 | 43.73 | 153259.0 | 31.5 |
| 990 | 2.2 | 0.205 | 0.031 | 0.207 | 0.002 | 45.0 | 22.0 | 0.004 | 2 | 12 | 6 | 2021-02-12 06:05:50.803695202 | 32.0 | 21.53 | 777.41 | 43.70 | 152841.0 | 31.5 |
| 991 | 2.2 | 0.205 | 0.031 | 0.207 | 0.002 | 45.0 | 22.0 | 0.004 | 2 | 12 | 6 | 2021-02-12 06:05:53.795462847 | 30.0 | 21.52 | 777.41 | 43.70 | 153399.0 | 30.2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1407742 | 0.6 | 0.001 | 0.009 | 0.011 | 0.035 | 44.0 | 17.0 | 0.002 | 4 | 1 | 23 | 2021-04-01 23:59:47.055900335 | 153.0 | 26.98 | 783.10 | 30.18 | 502865.0 | 153.1 |
| 1407743 | 0.6 | 0.001 | 0.009 | 0.011 | 0.035 | 44.0 | 17.0 | 0.002 | 4 | 1 | 23 | 2021-04-01 23:59:50.050112724 | 154.0 | 26.98 | 783.08 | 30.19 | 500990.0 | 154.0 |
| 1407744 | 0.6 | 0.001 | 0.009 | 0.011 | 0.035 | 44.0 | 17.0 | 0.002 | 4 | 1 | 23 | 2021-04-01 23:59:53.044603348 | 154.0 | 27.00 | 783.08 | 30.15 | 502865.0 | 154.0 |
| 1407745 | 0.6 | 0.001 | 0.009 | 0.011 | 0.035 | 44.0 | 17.0 | 0.002 | 4 | 1 | 23 | 2021-04-01 23:59:56.038767099 | 154.0 | 27.00 | 783.08 | 30.14 | 502865.0 | 154.0 |
| 1407746 | 0.6 | 0.001 | 0.009 | 0.011 | 0.035 | 44.0 | 17.0 | 0.002 | 4 | 1 | 23 | 2021-04-01 23:59:59.033584118 | 154.0 | 27.01 | 783.06 | 30.13 | 503242.0 | 153.9 |
1406760 rows × 18 columns
sinaica3.to_pickle("data/sinaica/sinaica-imputated.pickle.gz")
Hemos imputado con éxito todos los datos:
(sinaica2[sinaica2.delta > 2].
sort_values("delta", ascending=False)
[["Fecha", "delta"]].
reset_index(drop=True).
rename(columns={"delta": "Horas Faltantes"}).
head(10)
)
| Fecha | Horas Faltantes |
|---|
Reconocemos que tal vez no es el mejor método, pero requerimos más tiempo para explorar más sobre imputaciones en series de tiempo.