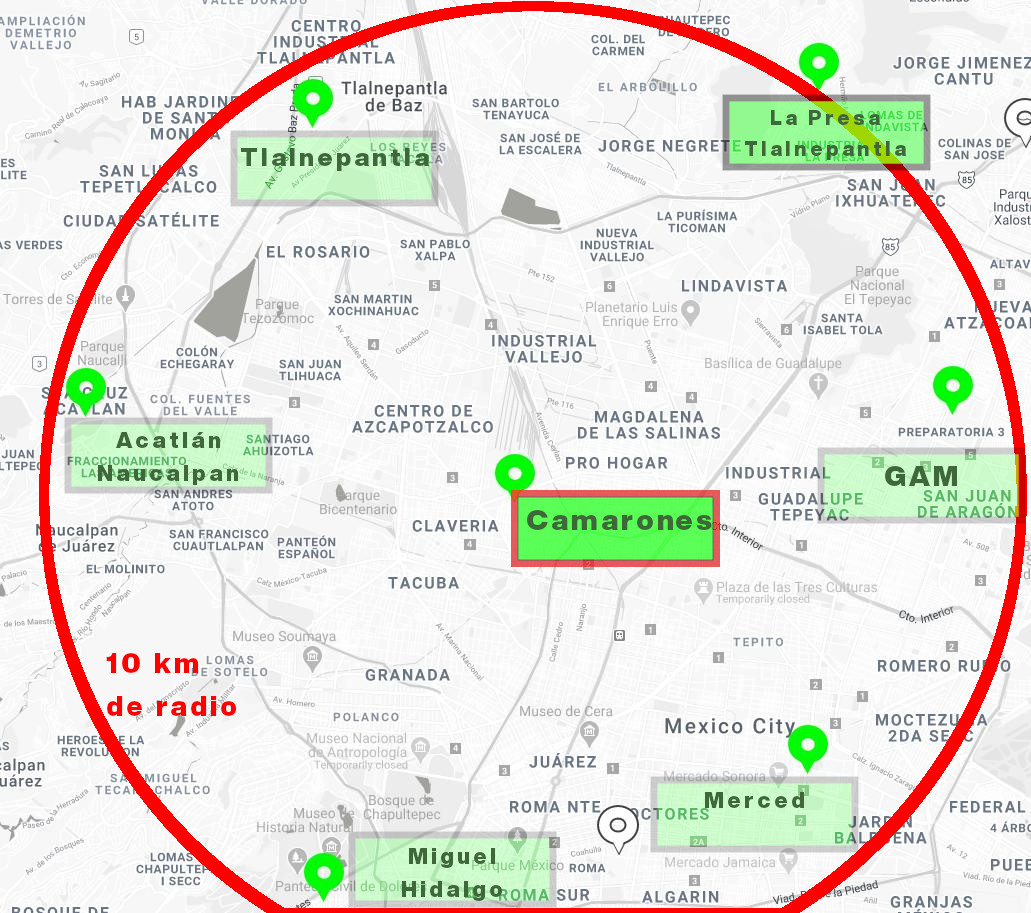
Nearby Air Quality Monitoring Stations¶
Here you may find the most proximate stations to "Camarones" which is the closest one to our sensor.
import os, gzip, json, re, stan, dplython, asyncio, nest_asyncio
#nest_asyncio.apply()
import warnings
from matplotlib import pyplot as plt
warnings.filterwarnings("ignore", category=DeprecationWarning)
from dplython import (DplyFrame, X, diamonds, select, sift,
sample_n, sample_frac, head, arrange, mutate, group_by,
summarize, DelayFunction, dfilter)
import seaborn as sns
from plotnine import *
from sklearn.linear_model import LinearRegression, Lasso
from sklearn.model_selection import train_test_split
from sklearn.metrics import (mean_squared_error,
r2_score,
mean_absolute_error)
import pandas as pd
import numpy as np
from IPython.display import display, Markdown, update_display
/home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject /home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject /home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject /home/jaa6766/.conda/envs/cuda/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
base_dir = "data/sinaica2/"
data_dir = os.path.join(
os.getcwd(),
base_dir
)
#df = pd.read_pickle("data/airdata/air.pickle")
display(Markdown(f"Listing data files from: {data_dir}..."))
generator = (file for file in os.listdir(data_dir)
if (re.match(r"Datos SINAICA - [-A-ZáéíóúÁÉÍÓÚa-z0-9. ]{30,80}\.csv", file) is not None))
num_files = len([j for (j, _) in enumerate(generator)]) + 1
generator.close()
generator = (file for file in os.listdir(data_dir)
if (re.match(r"Datos SINAICA - [-A-ZáéíóúÁÉÍÓÚa-z0-9. ]{30,80}\.csv", file) is not None))
sinaica = pd.read_pickle("data/sinaica.pickle")
display(Markdown(""), display_id="read_sinaica")
for (j, file_csv) in enumerate(generator):
##display(Markdown(f"{j+1}. File \"{file_csv}\""))
update_display(Markdown(f">Reading file {j+1} of {num_files} \"{file_csv}\""), display_id="read_sinaica")
##print("%d out of %d"%(j+1, num_files), end="")
try:
station, sensor = re.match(
r"Datos SINAICA - ([A-ZáéíóúÁÉÍÓÚa-z. ]{3,20}) - ([A-Z0-9a-z.]+) - [-0-9 ]+\.csv",
file_csv).groups()
#display(Markdown(f" * {station}, {sensor}"))
df2 = pd.read_csv(os.path.join(data_dir, file_csv))
df2 = df2.assign(Estacion=station)
sinaica = pd.concat([sinaica, df2])
#display(df2.head(3))
sinaica = sinaica.sort_values(by=["Fecha", "Estacion", "Parámetro"])
sinaica["Hora"] = sinaica[["Hora"]].replace("- .*$", "", regex=True)
##print(sinaica["Fecha"])
sinaica["Fecha"] = pd.to_datetime(sinaica["Fecha"])
##sinaica["Fecha"] = pd.to_datetime(sinaica["Fecha"] + " " + sinaica["Hora"])
sinaica.drop("Hora", axis=1, inplace=True)
sinaica = sinaica[(sinaica["Fecha"] >= "2021-01-01")].copy()
##display(Markdown(f"Done reading {j+1} files!"))
#display(pd.concat([sinaica.head(5), sinaica.tail(5)]))
except Exception as e:
print(f"\nError while reading file {file_csv}", type(e))
raise e
generator.close()
update_display(Markdown(f"Done!"), display_id="read_sinaica")
generator.close()
Listing data files from: /home/jaa6766/Documents/jorge3a/itam/deeplearning/dlfinal/data/sinaica2/...
Done!
sinaica
| Parámetro | Fecha | Valor | Unidad | Estacion | |
|---|---|---|---|---|---|
| 1 | CO | 2021-01-01 | 0.600 | ppm | Camarones |
| 1 | NO | 2021-01-01 | 0.006 | ppm | Camarones |
| 1 | NO2 | 2021-01-01 | 0.029 | ppm | Camarones |
| 1 | NOx | 2021-01-01 | 0.034 | ppm | Camarones |
| 1 | O3 | 2021-01-01 | 0.011 | ppm | Camarones |
| ... | ... | ... | ... | ... | ... |
| 34 | SO2 | 2021-10-08 | 0.002 | ppm | Merced |
| 35 | SO2 | 2021-10-08 | 0.001 | ppm | Merced |
| 36 | SO2 | 2021-10-08 | 0.001 | ppm | Merced |
| 37 | SO2 | 2021-10-08 | 0.000 | ppm | Merced |
| 38 | SO2 | 2021-10-08 | 0.001 | ppm | Merced |
196289 rows × 5 columns
Here you may find the most proximate stations to "Camarones" which is the closest one to our sensor.

(
ggplot(sinaica) +
geom_point(aes(x="Fecha", y="Valor", color="Parámetro")) +
facet_wrap("Parámetro", scales="free") +
labs(title="Visualization of the Pollulants") +
theme(axis_text_x=element_text(angle=90),
subplots_adjust={'wspace': 0.25, 'hspace': 0.25}
)
)
<ggplot: (8748608559861)>
(
ggplot(sinaica[(sinaica["Estacion"] == "Camarones")]) +
geom_point(aes(x="Fecha", y="Valor", color="Parámetro")) +
facet_wrap("Parámetro", scales="free") +
labs(title="Camarones Air Quality Monitoring Station") +
theme(axis_text_x=element_text(angle=90),
subplots_adjust={'wspace': 0.25, 'hspace': 0.25}
)
)
<ggplot: (8748605756781)>
sinaica.to_pickle("data/sinaica/sinaica.pickle")
Some of the missisng observatinos are caused by maintenance on the monitoring systems. So we could try to fill out the missing data with nearby government sesnsors. Then we propose to evaluate how the imputations work.
dsinaica = DplyFrame(sinaica)
dsinaica2 = (
dsinaica.
pivot_table(index=["Fecha", ], columns=["Estacion", "Parámetro"], values="Valor",
)
)
dsinaica2.columns = ["_".join(x).strip() for x in dsinaica2.columns]
dsinaica2.insert(0, "Fecha", dsinaica2.index)
dsinaica2.reset_index(drop=True, inplace=True)
dsinaica2.to_pickle("data/sinaica2/dsinaica.pickle")
dsinaica = DplyFrame(pd.read_pickle("data/sinaica2/dsinaica.pickle"))
dsinaica
| Fecha | Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | FES Acatlán_CO | ... | Miguel Hidalgo_O3 | Miguel Hidalgo_SO2 | Tlalnepantla_CO | Tlalnepantla_NO | Tlalnepantla_NO2 | Tlalnepantla_NOx | Tlalnepantla_O3 | Tlalnepantla_PM10 | Tlalnepantla_PM2.5 | Tlalnepantla_SO2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2021-01-01 00:00:00 | 0.600000 | 0.006000 | 0.029000 | 0.034 | 0.011000 | NaN | NaN | 0.002000 | 0.400000 | ... | 0.009 | 0.003 | 0.6 | NaN | 0.030 | 0.034 | 0.012 | 37.0 | 19.0 | 0.002 |
| 1 | 2021-01-01 01:00:00 | 1.000000 | 0.021000 | 0.038000 | 0.059 | 0.002000 | NaN | NaN | 0.002000 | 0.600000 | ... | 0.006 | 0.003 | 0.6 | NaN | 0.026 | 0.029 | 0.013 | 42.0 | 29.0 | 0.003 |
| 2 | 2021-01-01 02:00:00 | 0.800000 | 0.013000 | 0.035000 | 0.049 | 0.003000 | NaN | NaN | 0.001000 | 0.900000 | ... | 0.003 | 0.002 | 0.7 | NaN | 0.032 | 0.036 | 0.006 | 58.0 | 43.0 | 0.002 |
| 3 | 2021-01-01 03:00:00 | 1.000000 | 0.031000 | 0.034000 | 0.065 | 0.002000 | NaN | NaN | 0.001000 | 0.800000 | ... | 0.004 | 0.002 | 0.7 | NaN | 0.033 | 0.039 | 0.004 | 59.0 | 41.0 | 0.002 |
| 4 | 2021-01-01 04:00:00 | 0.600000 | 0.005000 | 0.029000 | 0.034 | 0.005000 | NaN | NaN | 0.001000 | 1.000000 | ... | 0.006 | 0.002 | 0.7 | NaN | 0.032 | 0.038 | 0.004 | 64.0 | 46.0 | 0.002 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2347 | 2021-10-04 00:00:00 | 0.441667 | 0.008292 | 0.015833 | NaN | 0.017167 | 22.173913 | 10.952381 | 0.000125 | 0.315000 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2348 | 2021-10-05 00:00:00 | 0.490000 | 0.010000 | 0.017000 | NaN | 0.013947 | 22.142857 | 8.736842 | 0.000000 | 0.466667 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2349 | 2021-10-06 00:00:00 | 0.542857 | 0.007571 | 0.022571 | NaN | 0.014333 | 25.150000 | 10.150000 | 0.000000 | 0.347619 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2350 | 2021-10-07 00:00:00 | 0.582609 | 0.011565 | 0.023130 | NaN | 0.021304 | 33.500000 | 15.428571 | 0.001783 | 0.447826 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2351 | 2021-10-08 00:00:00 | 0.738889 | 0.023778 | 0.026778 | NaN | 0.019667 | 41.266667 | 18.800000 | 0.010500 | 0.566667 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
2352 rows × 45 columns
dsinaica[dsinaica.isna().any(axis=1)]
| Fecha | Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | FES Acatlán_CO | ... | Miguel Hidalgo_O3 | Miguel Hidalgo_SO2 | Tlalnepantla_CO | Tlalnepantla_NO | Tlalnepantla_NO2 | Tlalnepantla_NOx | Tlalnepantla_O3 | Tlalnepantla_PM10 | Tlalnepantla_PM2.5 | Tlalnepantla_SO2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2021-01-01 00:00:00 | 0.600000 | 0.006000 | 0.029000 | 0.034 | 0.011000 | NaN | NaN | 0.002000 | 0.400000 | ... | 0.009 | 0.003 | 0.6 | NaN | 0.030 | 0.034 | 0.012 | 37.0 | 19.0 | 0.002 |
| 1 | 2021-01-01 01:00:00 | 1.000000 | 0.021000 | 0.038000 | 0.059 | 0.002000 | NaN | NaN | 0.002000 | 0.600000 | ... | 0.006 | 0.003 | 0.6 | NaN | 0.026 | 0.029 | 0.013 | 42.0 | 29.0 | 0.003 |
| 2 | 2021-01-01 02:00:00 | 0.800000 | 0.013000 | 0.035000 | 0.049 | 0.003000 | NaN | NaN | 0.001000 | 0.900000 | ... | 0.003 | 0.002 | 0.7 | NaN | 0.032 | 0.036 | 0.006 | 58.0 | 43.0 | 0.002 |
| 3 | 2021-01-01 03:00:00 | 1.000000 | 0.031000 | 0.034000 | 0.065 | 0.002000 | NaN | NaN | 0.001000 | 0.800000 | ... | 0.004 | 0.002 | 0.7 | NaN | 0.033 | 0.039 | 0.004 | 59.0 | 41.0 | 0.002 |
| 4 | 2021-01-01 04:00:00 | 0.600000 | 0.005000 | 0.029000 | 0.034 | 0.005000 | NaN | NaN | 0.001000 | 1.000000 | ... | 0.006 | 0.002 | 0.7 | NaN | 0.032 | 0.038 | 0.004 | 64.0 | 46.0 | 0.002 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2347 | 2021-10-04 00:00:00 | 0.441667 | 0.008292 | 0.015833 | NaN | 0.017167 | 22.173913 | 10.952381 | 0.000125 | 0.315000 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2348 | 2021-10-05 00:00:00 | 0.490000 | 0.010000 | 0.017000 | NaN | 0.013947 | 22.142857 | 8.736842 | 0.000000 | 0.466667 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2349 | 2021-10-06 00:00:00 | 0.542857 | 0.007571 | 0.022571 | NaN | 0.014333 | 25.150000 | 10.150000 | 0.000000 | 0.347619 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2350 | 2021-10-07 00:00:00 | 0.582609 | 0.011565 | 0.023130 | NaN | 0.021304 | 33.500000 | 15.428571 | 0.001783 | 0.447826 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2351 | 2021-10-08 00:00:00 | 0.738889 | 0.023778 | 0.026778 | NaN | 0.019667 | 41.266667 | 18.800000 | 0.010500 | 0.566667 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
1694 rows × 45 columns
We can tell that "Camarones", the closest one, has missing data on all variables.
camarones = dsinaica[dsinaica.filter(like="Camarones").isna().any(axis=1)].filter(like="Camarones")
camarones
| Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.600000 | 0.006000 | 0.029000 | 0.034 | 0.011000 | NaN | NaN | 0.002000 |
| 1 | 1.000000 | 0.021000 | 0.038000 | 0.059 | 0.002000 | NaN | NaN | 0.002000 |
| 2 | 0.800000 | 0.013000 | 0.035000 | 0.049 | 0.003000 | NaN | NaN | 0.001000 |
| 3 | 1.000000 | 0.031000 | 0.034000 | 0.065 | 0.002000 | NaN | NaN | 0.001000 |
| 4 | 0.600000 | 0.005000 | 0.029000 | 0.034 | 0.005000 | NaN | NaN | 0.001000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2347 | 0.441667 | 0.008292 | 0.015833 | NaN | 0.017167 | 22.173913 | 10.952381 | 0.000125 |
| 2348 | 0.490000 | 0.010000 | 0.017000 | NaN | 0.013947 | 22.142857 | 8.736842 | 0.000000 |
| 2349 | 0.542857 | 0.007571 | 0.022571 | NaN | 0.014333 | 25.150000 | 10.150000 | 0.000000 |
| 2350 | 0.582609 | 0.011565 | 0.023130 | NaN | 0.021304 | 33.500000 | 15.428571 | 0.001783 |
| 2351 | 0.738889 | 0.023778 | 0.026778 | NaN | 0.019667 | 41.266667 | 18.800000 | 0.010500 |
791 rows × 8 columns
display(Markdown(f"Then we can look forward to avoid loosing a big portion of data: " +
f'{100*dsinaica[dsinaica.filter(like="Camarones").isna().any(axis=1)].filter(like="Camarones").shape[0]/dsinaica.shape[0]:.2f}%' +
f' by using imputation. Our goal is to evaluate the different imputation methods in order to have data to back our decision.'
))
Then we can look forward to avoid loosing a big portion of data: 33.63% by using imputation. Our goal is to evaluate the different imputation methods in order to have data to back our decision.
dsinaica[~dsinaica.filter(like="Camarones").isna().any(axis=1)].filter(like="Camarones")
| Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | |
|---|---|---|---|---|---|---|---|---|
| 478 | 0.933333 | 0.035333 | 0.03275 | 0.028 | 0.0195 | 43.047619 | 21.333333 | 0.004708 |
| 480 | 0.500000 | 0.021000 | 0.02400 | 0.046 | 0.0020 | 30.000000 | 14.000000 | 0.008000 |
| 481 | 0.600000 | 0.017000 | 0.02300 | 0.039 | 0.0020 | 28.000000 | 16.000000 | 0.006000 |
| 482 | 0.500000 | 0.023000 | 0.02200 | 0.046 | 0.0020 | 38.000000 | 26.000000 | 0.005000 |
| 483 | 0.600000 | 0.030000 | 0.02100 | 0.051 | 0.0020 | 35.000000 | 21.000000 | 0.003000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2167 | 0.400000 | 0.003000 | 0.01100 | 0.013 | 0.0160 | 69.000000 | 7.000000 | 0.001000 |
| 2168 | 0.400000 | 0.002000 | 0.01100 | 0.012 | 0.0180 | 71.000000 | 9.000000 | 0.001000 |
| 2169 | 0.400000 | 0.002000 | 0.01300 | 0.015 | 0.0160 | 37.000000 | 9.000000 | 0.001000 |
| 2170 | 0.400000 | 0.002000 | 0.01900 | 0.021 | 0.0120 | 19.000000 | 0.000000 | 0.001000 |
| 2171 | 0.400000 | 0.001000 | 0.01400 | 0.015 | 0.0210 | 61.000000 | 21.000000 | 0.001000 |
1561 rows × 8 columns
camarones_full = dsinaica[~dsinaica.filter(like="Camarones").isna().any(axis=1)]
camarones_train, camarones_test = train_test_split(camarones_full,
test_size=0.3, random_state=175904,
shuffle=False
)
camarones_val = dsinaica[dsinaica.filter(like="Camarones").isna().any(axis=1)]
display(Markdown(f"* Complete observations: {camarones_full.shape[0]} (100%)."))
display(Markdown(f">* Complete observations on Training Set: {camarones_train.shape[0]} (~70%)."))
display(Markdown(f">* Complete observations on Test Set: {camarones_test.shape[0]} (~30%)."))
display(Markdown(f"* Incomplete Observations: {camarones_val.shape[0]}."))
- Complete observations on Training Set: 1092 (~70%).
- Complete observations on Test Set: 469 (~30%).
dsinaica[~dsinaica.filter(like="PM10").isna().any(axis=1)].filter(like="PM10")
| Camarones_PM10 | FES Acatlán_PM10 | Gustavo A. Madero_PM10 | Merced_PM10 | Tlalnepantla_PM10 | |
|---|---|---|---|---|---|
| 478 | 43.047619 | 34.0 | 32.0 | 39.727273 | 23.0 |
| 480 | 30.000000 | 15.0 | 33.0 | 34.000000 | 32.0 |
| 481 | 28.000000 | 11.0 | 32.0 | 29.000000 | 24.0 |
| 482 | 38.000000 | 15.0 | 28.0 | 33.000000 | 41.0 |
| 483 | 35.000000 | 15.0 | 25.0 | 32.000000 | 20.0 |
| ... | ... | ... | ... | ... | ... |
| 2166 | 71.000000 | 178.0 | 69.0 | 49.000000 | 90.0 |
| 2167 | 69.000000 | 162.0 | 33.0 | 36.000000 | 52.0 |
| 2168 | 71.000000 | 49.0 | 32.0 | 24.000000 | 22.0 |
| 2170 | 19.000000 | 14.0 | 19.0 | 21.000000 | 11.0 |
| 2171 | 61.000000 | 56.0 | 44.0 | 44.000000 | 55.0 |
1305 rows × 5 columns
def describe_with_na(dataframe):
dna = dataframe.isna()
dna = dna.astype('int').sum()
dna.name = "NAs"
dataframe = dataframe.describe()
dataframe = dataframe.append(dna)
dataframe = dataframe.T
dataframe.insert(0, "Estacion", dataframe.index)
return dataframe
describe_with_na(camarones_train.filter(regex=r"PM10", axis=1))
| Estacion | count | mean | std | min | 25% | 50% | 75% | max | NAs | |
|---|---|---|---|---|---|---|---|---|---|---|
| Camarones_PM10 | Camarones_PM10 | 1092.0 | 58.163765 | 27.817189 | 0.0 | 42.0 | 55.0 | 71.0 | 437.0 | 0.0 |
| FES Acatlán_PM10 | FES Acatlán_PM10 | 1011.0 | 48.669366 | 32.624587 | 0.0 | 29.0 | 43.0 | 61.0 | 388.0 | 81.0 |
| Gustavo A. Madero_PM10 | Gustavo A. Madero_PM10 | 1029.0 | 52.906706 | 26.733354 | 0.0 | 34.0 | 50.0 | 67.0 | 352.0 | 63.0 |
| Merced_PM10 | Merced_PM10 | 1084.0 | 56.798977 | 21.410434 | 5.0 | 42.0 | 55.0 | 69.0 | 184.0 | 8.0 |
| Tlalnepantla_PM10 | Tlalnepantla_PM10 | 1034.0 | 52.254352 | 29.446065 | 0.0 | 36.0 | 48.0 | 63.0 | 416.0 | 58.0 |
describe_with_na(dsinaica.filter(regex=r"Camarones", axis=1))
| Estacion | count | mean | std | min | 25% | 50% | 75% | max | NAs | |
|---|---|---|---|---|---|---|---|---|---|---|
| Camarones_CO | Camarones_CO | 2241.0 | 0.767037 | 0.412628 | 0.000000 | 0.500000 | 0.700 | 0.900 | 3.200 | 111.0 |
| Camarones_NO | Camarones_NO | 2227.0 | 0.024599 | 0.043639 | 0.000000 | 0.003000 | 0.007 | 0.026 | 0.432 | 125.0 |
| Camarones_NO2 | Camarones_NO2 | 2227.0 | 0.031139 | 0.015144 | 0.003000 | 0.020000 | 0.029 | 0.040 | 0.111 | 125.0 |
| Camarones_NOx | Camarones_NOx | 2042.0 | 0.056961 | 0.054418 | 0.004000 | 0.022000 | 0.039 | 0.071 | 0.499 | 310.0 |
| Camarones_O3 | Camarones_O3 | 2233.0 | 0.026094 | 0.022991 | 0.001000 | 0.005000 | 0.021 | 0.039 | 0.103 | 119.0 |
| Camarones_PM10 | Camarones_PM10 | 1749.0 | 56.638261 | 26.729443 | 0.000000 | 40.000000 | 54.000 | 70.000 | 437.000 | 603.0 |
| Camarones_PM2.5 | Camarones_PM2.5 | 1765.0 | 24.872841 | 12.620210 | 0.000000 | 16.000000 | 24.000 | 32.000 | 126.000 | 587.0 |
| Camarones_SO2 | Camarones_SO2 | 2190.0 | 0.006284 | 0.012464 | -0.000048 | 0.001217 | 0.003 | 0.005 | 0.162 | 162.0 |
contaminante = "Camarones_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "FES Acatlán_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "Gustavo A. Madero_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "Tlalnepantla_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
contaminante = "Merced_PM10"
plt.boxplot(x=camarones_train[~camarones_train.filter(regex=contaminante, axis=1).isna().any(axis=1)][contaminante])
plt.xlabel(contaminante)
plt.show()
(
ggplot(camarones_train[["Camarones_PM10"]]) +
geom_histogram(aes(x="Camarones_PM10"), bins=100) +
geom_vline(aes(xintercept=camarones_train["Camarones_PM10"].mean()),
color="red") +
labs(title="Histogram of PM10 in Camarones.")
)
<ggplot: (8748605798857)>
(
ggplot(camarones_train[["Gustavo A. Madero_PM10"]]) +
geom_histogram(aes(x="Gustavo A. Madero_PM10"), bins=100) +
geom_vline(aes(xintercept=camarones_train["Gustavo A. Madero_PM10"].mean()),
color="red") +
labs(title="Histogram of PM10 in Gustavo A. Madero.")
)
/home/jaa6766/.conda/envs/cuda/lib/python3.7/site-packages/plotnine/layer.py:372: PlotnineWarning: stat_bin : Removed 63 rows containing non-finite values.
<ggplot: (8748605795749)>
(
ggplot(camarones_train[["Merced_PM10"]]) +
geom_histogram(aes(x="Merced_PM10"), bins=100) +
labs(title="Histogram of PM10 in La Merced.")
)
/home/jaa6766/.conda/envs/cuda/lib/python3.7/site-packages/plotnine/layer.py:372: PlotnineWarning: stat_bin : Removed 8 rows containing non-finite values.
<ggplot: (8748605840569)>
(
ggplot(camarones_train[["FES Acatlán_PM10"]]) +
geom_histogram(aes(x="FES Acatlán_PM10"), bins=100) +
labs(title="Histogram of PM10 in FES Acatlán.")
)
/home/jaa6766/.conda/envs/cuda/lib/python3.7/site-packages/plotnine/layer.py:372: PlotnineWarning: stat_bin : Removed 81 rows containing non-finite values.
<ggplot: (8748598829781)>
(
ggplot(camarones_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]) +
geom_point(aes(x="Gustavo A. Madero_PM10", y="Camarones_PM10")) +
labs(title="Scatter Plot for PM10 for GAM and Camarones.")
)
<ggplot: (8748598907869)>
(
ggplot(camarones_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]) +
geom_point(aes(x="Merced_PM10", y="Camarones_PM10")) +
labs(title="Scatter Plot for PM10 for Camarones and Merced.")
)
<ggplot: (8748598840809)>
corr = camarones_train.filter(like="PM10", axis=1)[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)].corr()
corr
| Camarones_PM10 | FES Acatlán_PM10 | Gustavo A. Madero_PM10 | Merced_PM10 | Tlalnepantla_PM10 | |
|---|---|---|---|---|---|
| Camarones_PM10 | 1.000000 | 0.588032 | 0.573777 | 0.642624 | 0.633015 |
| FES Acatlán_PM10 | 0.588032 | 1.000000 | 0.561526 | 0.596102 | 0.755112 |
| Gustavo A. Madero_PM10 | 0.573777 | 0.561526 | 1.000000 | 0.779445 | 0.588039 |
| Merced_PM10 | 0.642624 | 0.596102 | 0.779445 | 1.000000 | 0.599155 |
| Tlalnepantla_PM10 | 0.633015 | 0.755112 | 0.588039 | 0.599155 | 1.000000 |
#corr = corr.loc[:, ["PM2.5", "PM10"]]
mask = None
#mask = np.triu(np.ones_like(corr*-1, dtype=bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
#cmap = sns.palettes.blend_palette(["#ffffff", "#000055"],
# n_colors=16,
# as_cmap=True)
cmap = sns.color_palette("light:blue", as_cmap=True)
plt.title("Correlations Heat Map: " +
"(Absolute Values of Correlations).", loc='left')
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr, mask=mask, cmap=cmap, #vmax=.3, center=0,
square=True, linewidths=.5, cbar_kws={"shrink": .5})
#plt.xlabel("Variables Respuesta")
#plt.ylabel("Variables Predictoras")
plt.show()
corr = camarones_train.filter(like="Camarones", axis=1)[~camarones_train.filter(regex=r"Camarones", axis=1).isna().any(axis=1)].corr()
corr
| Camarones_CO | Camarones_NO | Camarones_NO2 | Camarones_NOx | Camarones_O3 | Camarones_PM10 | Camarones_PM2.5 | Camarones_SO2 | |
|---|---|---|---|---|---|---|---|---|
| Camarones_CO | 1.000000 | 0.745593 | 0.771172 | 0.839178 | -0.455741 | 0.297277 | 0.425596 | 0.109512 |
| Camarones_NO | 0.745593 | 1.000000 | 0.456594 | 0.965661 | -0.440542 | 0.124081 | 0.200851 | 0.105557 |
| Camarones_NO2 | 0.771172 | 0.456594 | 1.000000 | 0.656534 | -0.554310 | 0.279483 | 0.435905 | 0.160720 |
| Camarones_NOx | 0.839178 | 0.965661 | 0.656534 | 1.000000 | -0.522438 | 0.181161 | 0.288952 | 0.133070 |
| Camarones_O3 | -0.455741 | -0.440542 | -0.554310 | -0.522438 | 1.000000 | 0.030864 | 0.003132 | -0.192407 |
| Camarones_PM10 | 0.297277 | 0.124081 | 0.279483 | 0.181161 | 0.030864 | 1.000000 | 0.610515 | 0.157361 |
| Camarones_PM2.5 | 0.425596 | 0.200851 | 0.435905 | 0.288952 | 0.003132 | 0.610515 | 1.000000 | 0.298078 |
| Camarones_SO2 | 0.109512 | 0.105557 | 0.160720 | 0.133070 | -0.192407 | 0.157361 | 0.298078 | 1.000000 |
#corr = corr.loc[:, ["PM2.5", "PM10"]]
mask = None
#mask = np.triu(np.ones_like(corr*-1, dtype=bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
#cmap = sns.palettes.blend_palette(["#ffffff", "#000055"],
# n_colors=16,
# as_cmap=True)
#cmap = sns.diverging_palette(250, 20, as_cmap=True)
cmap = sns.color_palette("coolwarm", as_cmap=True)
plt.title("Correlations Heat Map: " +
"(Absolute Values of Correlations).", loc='left')
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr, mask=mask, cmap=cmap, #vmax=.3, center=0,
square=True, linewidths=.5, cbar_kws={"shrink": .5})
#plt.xlabel("Variables Respuesta")
#plt.ylabel("Variables Predictoras")
plt.show()
(
ggplot(camarones_train[["Camarones_PM2.5"]]) +
geom_histogram(aes(x="Camarones_PM2.5"), bins=100) +
labs(title="Histogram for PM2.5 for Camarones.")
)
<ggplot: (8748609230585)>
Removemos observaciones incompletas para realizar la regresión.
Y_train = camarones_train["Camarones_PM10"]
Y_test = camarones_test["Camarones_PM10"]
X_train = camarones_train.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
X_test = camarones_test.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
X_train = camarones_train.filter(regex="(Merced|Tlalnepantla)")
X_test = camarones_test.filter(regex="(Merced|Tlalnepantla)")
## guardar los parquet para correr Stan en R
camarones_train.to_parquet("data/camarones_train.parquet")
camarones_test.to_parquet("data/camarones_test.parquet")
### Código para filtrar solo las columnas con PM10
### Es decir, estaciones que tienen el contaminante PM10
X_test = X_test.filter(regex=r"PM10", axis=1)
X_train = X_train.filter(regex=r"PM10", axis=1)
## Removemos observaciones incompletas
X_train = X_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
X_test = X_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_train = Y_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_test = Y_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
reg = LinearRegression(n_jobs=8).fit(X_train, Y_train)
Yhat_train_lin = reg.predict(X_train)
Yhat_test_lin = reg.predict(X_test)
X_train.columns
Index(['Merced_PM10', 'Tlalnepantla_PM10'], dtype='object')
reg.coef_
array([0.53026139, 0.36873538])
reg.intercept_
8.861413306258605
r2_score(Y_train, Yhat_train_lin)
0.5089006183870153
r2_score(Y_test, Yhat_test_lin)
-0.4162199659052408
mean_squared_error(Y_train, Yhat_train_lin)
371.5820988359095
mean_squared_error(Y_test, Yhat_test_lin)
845.7794576326868
mean_absolute_error(Y_train, Yhat_train_lin)
10.931473014049846
mean_absolute_error(Y_test, Yhat_test_lin)
13.047752185462492
(
ggplot(pd.DataFrame({
"Y_train": Y_train,
"Yhat_train": Yhat_train_lin
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8748609122933)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_lin
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8748608796597)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_lin
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8748608756745)>
Y_train = camarones_train["Camarones_PM10"]
Y_test = camarones_test["Camarones_PM10"]
X_train = camarones_train.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
X_test = camarones_test.filter(regex=r"^(?!Camarones_PM10|Fecha)", axis=1)
### Código para filtrar solo las columnas con PM10
### Es decir, estaciones que tienen el contaminante PM10
X_test = X_test.filter(regex=r"PM10", axis=1)
X_train = X_train.filter(regex=r"PM10", axis=1)
## Removemos observaciones incompletas
X_train = X_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
X_test = X_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_train = Y_train[~camarones_train.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
Y_test = Y_test[~camarones_test.filter(regex=r"PM10", axis=1).isna().any(axis=1)]
reg = Lasso().fit(X_train, Y_train)
Yhat_train_lasso = reg.predict(X_train)
Yhat_test_lasso = reg.predict(X_test)
reg.coef_
array([0.10494418, 0.0689564 , 0.43803925, 0.28309869])
reg.intercept_
9.793830115973797
r2_score(Y_train, Yhat_train_lasso)
0.5169119069776114
r2_score(Y_test, Yhat_test_lasso)
-0.11224862731990415
mean_squared_error(Y_train, Yhat_train_lasso)
365.52049187746337
mean_squared_error(Y_test, Yhat_test_lasso)
664.2450067183082
mean_absolute_error(Y_train, Yhat_train_lasso)
10.791035566246128
mean_absolute_error(Y_test, Yhat_test_lasso)
12.489847967051261
(
ggplot(pd.DataFrame({
"Y_train": Y_train,
"Yhat_train": Yhat_train_lasso
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8748608657061)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_lasso
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8748608729533)>
Yhat_train_mean = X_train.mean(axis=1)
Yhat_test_mean = X_test.mean(axis=1)
mean_squared_error(Y_train, Yhat_train_mean)
422.1441318590655
mean_squared_error(Y_test, Yhat_test_mean)
579.0378415931376
mean_absolute_error(Y_train, Yhat_train_mean)
12.22969669074124
mean_absolute_error(Y_test, Yhat_test_mean)
13.812958507896164
(
ggplot(pd.DataFrame({
"Y_train": Y_train,
"Yhat_train": Yhat_train_mean
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"),
color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"),
color='red', alpha=.5)
)
<ggplot: (8748608608917)>
(
ggplot(pd.DataFrame({
"Y_train": Y_test,
"Yhat_train": Yhat_test_mean
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"),
color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_train"),
color='red', alpha=.5)
)
<ggplot: (8748608547589)>
from sklearn.linear_model import PoissonRegressor
reg = PoissonRegressor(alpha=1e3)
reg_fit = reg.fit(X_train, Y_train)
Yhat_train_glm = reg_fit.predict(X_train)
(Yhat_train_glm.shape, Y_train.shape)
((886,), (886,))
Yhat_test_glm = reg_fit.predict(X_test)
(Yhat_test_glm.shape, Y_test.shape)
((367,), (367,))
mean_squared_error(Yhat_train_glm
, Y_train)
562.8146939425658
mean_squared_error(Yhat_test_glm, Y_test)
7875.849124574941
mean_absolute_error(Yhat_train_glm, Y_train)
12.15806111211628
mean_absolute_error(Yhat_test_glm, Y_test)
17.957714790509712
(
ggplot(pd.DataFrame({
"Y_train": Y_train.reset_index(drop=True),
"Yhat_train": Yhat_train_glm
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8748609151741)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_glm
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8748608529529)>
from sklearn.neighbors import KNeighborsRegressor
knn = KNeighborsRegressor(n_neighbors=20, p=1)
knn_fit = knn.fit(X_train, Y_train)
Yhat_train_knn = knn_fit.predict(X_train)
(Yhat_train_knn.shape, Y_train.shape)
((886,), (886,))
Yhat_test_knn = knn_fit.predict(X_test)
(Yhat_test_knn.shape, Y_test.shape)
((367,), (367,))
mean_squared_error(Yhat_train_knn,
Y_train)
347.29259904240126
mean_squared_error(Yhat_test_knn, Y_test)
325.37804270928683
mean_absolute_error(Yhat_train_knn, Y_train)
10.748652093480683
mean_absolute_error(Yhat_test_knn, Y_test)
11.676590585214306
(
ggplot(pd.DataFrame({
"Y_train": Y_train.reset_index(drop=True),
"Yhat_train": Yhat_train_knn
})) +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Y_train"), color='blue') +
geom_point(aes(x=range(0, Y_train.shape[0]), y="Yhat_train"), color='red')
)
<ggplot: (8748608731669)>
(
ggplot(pd.DataFrame({
"Y_test": Y_test,
"Yhat_test": Yhat_test_knn
})) +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Y_test"), color='blue') +
geom_point(aes(x=range(0, Y_test.shape[0]), y="Yhat_test"), color='red')
)
<ggplot: (8748608731681)>
df = pd.DataFrame({
"Y_test": Y_test.reset_index(drop=True),
#"Yhat_test": Yhat_test_stan
})
df = df[~df.isnull().any(axis=1)]
df2 = pd.DataFrame({
"Model": [
"1.2 Regresión Lineal",
"1.3 Lasso",
#"1.4 Stan",
"1.5 Media",
"1.6 GLM",
"1.7 KNN"
],
"MSE (Train Set)": [
mean_squared_error(Y_train, Yhat_train_lin),
mean_squared_error(Y_train, Yhat_train_lasso),
#mean_squared_error(Y_train, Yhat_train_stan),
mean_squared_error(Y_train, Yhat_train_mean),
mean_squared_error(Y_train, Yhat_train_glm),
mean_squared_error(Y_train, Yhat_train_knn),
],
"MAE (Train Set)": [
mean_absolute_error(Y_train, Yhat_train_lin),
mean_absolute_error(Y_train, Yhat_train_lasso),
#mean_absolute_error(Y_train, Yhat_train_stan),
mean_absolute_error(Y_train, Yhat_train_mean),
mean_absolute_error(Y_train, Yhat_train_glm),
mean_absolute_error(Y_train, Yhat_train_knn),
],
"MSE (Test Set)": [
mean_squared_error(Y_test, Yhat_test_lin),
mean_squared_error(Y_test, Yhat_test_lasso),
#mean_squared_error(df[["Y_test"]], df[["Yhat_test"]]),
mean_squared_error(Y_test, Yhat_test_mean),
mean_squared_error(Y_test, Yhat_test_glm),
mean_squared_error(Y_test, Yhat_test_knn),
],
"MAE (Test Set)": [
mean_absolute_error(Y_test, Yhat_test_lin),
mean_absolute_error(Y_test, Yhat_test_lasso),
#mean_absolute_error(df[["Y_test"]], df[["Yhat_test"]]),
mean_absolute_error(Y_test, Yhat_test_mean),
mean_absolute_error(Y_test, Yhat_test_glm),
mean_absolute_error(Y_test, Yhat_test_knn),
]
})
df2.sort_values(by="MSE (Test Set)")
| Model | MSE (Train Set) | MAE (Train Set) | MSE (Test Set) | MAE (Test Set) | |
|---|---|---|---|---|---|
| 4 | 1.7 KNN | 347.292599 | 10.748652 | 325.378043 | 11.676591 |
| 2 | 1.5 Media | 422.144132 | 12.229697 | 579.037842 | 13.812959 |
| 1 | 1.3 Lasso | 365.520492 | 10.791036 | 664.245007 | 12.489848 |
| 0 | 1.2 Regresión Lineal | 371.582099 | 10.931473 | 845.779458 | 13.047752 |
| 3 | 1.6 GLM | 562.814694 | 12.158061 | 7875.849125 | 17.957715 |
Given that we all further treatment to use the data should be in a sequential fashion, ie as timeseries: we found that linear interpolation is adequate.
Then in the next section we are detailing it.
We found in the EDA and in previous sections that Merced has similar data as Camarones, and it has fewer incomplete observations (missing data).
merced = dsinaica.filter(regex='^Merced_').describe()
dsinaica.shape[0] - merced.iloc[0]
Merced_CO 70.0 Merced_NO 971.0 Merced_NO2 88.0 Merced_NOx 88.0 Merced_O3 82.0 Merced_PM10 31.0 Merced_PM2.5 36.0 Merced_SO2 64.0 Name: count, dtype: float64
Camarones has more incomplete observations:
camarones = dsinaica.filter(regex='^Camarones_').describe()
dsinaica.shape[0] - camarones.iloc[0]
Camarones_CO 111.0 Camarones_NO 125.0 Camarones_NO2 125.0 Camarones_NOx 310.0 Camarones_O3 119.0 Camarones_PM10 603.0 Camarones_PM2.5 587.0 Camarones_SO2 162.0 Name: count, dtype: float64
Using those results we create a new dataframe with those imputations.
The data in the dataframe have the following columns (vars) in the following manner:
CO: Merced
NO: Camarones
NO2: Merced
NOx: Merced
O3: Merced
PM10: Merced
PM2.5: Merced
We used a lag to use the missing hours and found these gaps in the time line:
sinaica_imputated = pd.DataFrame(dsinaica["Fecha"])
sinaica_imputated["CO"] = dsinaica["Merced_CO"]
sinaica_imputated["NO"] = dsinaica["Camarones_NO"]
sinaica_imputated["NO2"] = dsinaica["Merced_NO2"]
sinaica_imputated["NOx"] = dsinaica["Merced_NOx"]
sinaica_imputated["O3"] = dsinaica["Merced_O3"]
sinaica_imputated["PM10"] = dsinaica["Merced_PM10"]
sinaica_imputated["PM2.5"] = dsinaica["Merced_PM2.5"]
sinaica_imputated["SO2"] = dsinaica["Merced_SO2"]
sinaica_imputated["datetime"] = pd.to_datetime(sinaica_imputated["Fecha"])
sinaica_imputated = sinaica_imputated[~sinaica_imputated.isna().any(axis=1)]
sinaica_imputated["year"] = [dt.year for dt in sinaica_imputated.datetime]
sinaica_imputated["month"] = [dt.month for dt in sinaica_imputated.datetime]
sinaica_imputated["day"] = [dt.day for dt in sinaica_imputated.datetime]
sinaica_imputated["hour"] = [dt.hour for dt in sinaica_imputated.datetime]
sinaica_imputated["datetime-1"] = sinaica_imputated["datetime"].shift(1)
sinaica_imputated["delta"] = sinaica_imputated["datetime"] - sinaica_imputated["datetime-1"]
sinaica_imputated["delta"] = (sinaica_imputated["delta"].dt.seconds // 60**2)
sinaica_imputated["imputated"] = False
sinaica_imputated = sinaica_imputated[~sinaica_imputated.isna().any(axis=1)]
sinaica_imputated.reset_index(drop=True, inplace=True)
sinaica_imputated
| Fecha | CO | NO | NO2 | NOx | O3 | PM10 | PM2.5 | SO2 | datetime | year | month | day | hour | datetime-1 | delta | imputated | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2021-01-01 02:00:00 | 1.100000 | 0.013000 | 0.032000 | 0.039000 | 0.004000 | 37.000000 | 24.000000 | 0.003000 | 2021-01-01 02:00:00 | 2021 | 1 | 1 | 2 | 2021-01-01 00:00:00 | 2.0 | False |
| 1 | 2021-01-01 03:00:00 | 1.200000 | 0.031000 | 0.033000 | 0.043000 | 0.001000 | 49.000000 | 39.000000 | 0.003000 | 2021-01-01 03:00:00 | 2021 | 1 | 1 | 3 | 2021-01-01 02:00:00 | 1.0 | False |
| 2 | 2021-01-01 04:00:00 | 1.200000 | 0.005000 | 0.031000 | 0.039000 | 0.002000 | 80.000000 | 65.000000 | 0.003000 | 2021-01-01 04:00:00 | 2021 | 1 | 1 | 4 | 2021-01-01 03:00:00 | 1.0 | False |
| 3 | 2021-01-01 05:00:00 | 1.200000 | 0.016000 | 0.028000 | 0.036000 | 0.002000 | 89.000000 | 75.000000 | 0.003000 | 2021-01-01 05:00:00 | 2021 | 1 | 1 | 5 | 2021-01-01 04:00:00 | 1.0 | False |
| 4 | 2021-01-01 06:00:00 | 1.400000 | 0.024000 | 0.029000 | 0.060000 | 0.001000 | 75.000000 | 64.000000 | 0.003000 | 2021-01-01 06:00:00 | 2021 | 1 | 1 | 6 | 2021-01-01 05:00:00 | 1.0 | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2129 | 2021-10-04 00:00:00 | 0.545833 | 0.008292 | 0.019083 | 0.026875 | 0.015833 | 11.826087 | 7.913043 | 0.000750 | 2021-10-04 00:00:00 | 2021 | 10 | 4 | 0 | 2021-10-03 00:00:00 | 0.0 | False |
| 2130 | 2021-10-05 00:00:00 | 0.563158 | 0.010000 | 0.019722 | 0.030500 | 0.012278 | 11.090909 | 6.772727 | 0.000556 | 2021-10-05 00:00:00 | 2021 | 10 | 5 | 0 | 2021-10-04 00:00:00 | 0.0 | False |
| 2131 | 2021-10-06 00:00:00 | 0.672222 | 0.007571 | 0.026111 | 0.035611 | 0.011000 | 18.722222 | 11.833333 | 0.000111 | 2021-10-06 00:00:00 | 2021 | 10 | 6 | 0 | 2021-10-05 00:00:00 | 0.0 | False |
| 2132 | 2021-10-07 00:00:00 | 0.713636 | 0.011565 | 0.028636 | 0.040318 | 0.017909 | 26.772727 | 17.000000 | 0.001045 | 2021-10-07 00:00:00 | 2021 | 10 | 7 | 0 | 2021-10-06 00:00:00 | 0.0 | False |
| 2133 | 2021-10-08 00:00:00 | 0.758824 | 0.023778 | 0.029412 | 0.050588 | 0.017941 | 29.000000 | 17.705882 | 0.008176 | 2021-10-08 00:00:00 | 2021 | 10 | 8 | 0 | 2021-10-07 00:00:00 | 0.0 | False |
2134 rows × 17 columns
These are the missing gaps:
(sinaica_imputated[sinaica_imputated.delta > 2].
sort_values("delta", ascending=False)
[["Fecha", "delta"]].
reset_index(drop=True).
rename(columns={"delta": "Missing observations",
"Fecha": "Date"
}).
head(10)
)
| Date | Missing observations | |
|---|---|---|
| 0 | 2021-01-09 13:00:00 | 13.0 |
| 1 | 2021-03-11 23:00:00 | 12.0 |
| 2 | 2021-03-17 09:00:00 | 9.0 |
| 3 | 2021-03-17 00:00:00 | 8.0 |
| 4 | 2021-01-21 23:00:00 | 8.0 |
| 5 | 2021-02-26 08:00:00 | 8.0 |
| 6 | 2021-02-26 00:00:00 | 7.0 |
| 7 | 2021-02-22 15:00:00 | 6.0 |
| 8 | 2021-02-26 14:00:00 | 6.0 |
| 9 | 2021-01-01 18:00:00 | 5.0 |
Realizamos una interpolación quedando los datos así:
%%time
def interpolate_missing(df, idx, hours=1):
"""
Function to interpolate missing.
examples
airdata2 = interpolate_missing(airdata, idx = airdata[airdata.delta > 100].index[0])
airdata2 = interpolate_missing(airdata, 194044)
"""
np.random.seed(175904)
df_prev = df.loc[idx - 1]
a = df_prev
b = df.loc[idx]
offsetEnd = pd.offsets.Hour(1) # remove closed form
out = {}
out["datetime"] = pd.date_range(a["datetime"] + offsetEnd,
b["datetime"] - offsetEnd,
freq='1h')
out["datetime"] = out["datetime"].set_names("datetime")
for v in ['CO', 'NO', 'NO2', 'NOx', 'O3', 'PM10', 'PM2.5', 'SO2']:
i = [i+1 for i, d in enumerate(out["datetime"])]
m = (b[v] - a[v])/len(out["datetime"])
sd = 0.7*np.std(df_prev[v])
rnds = np.random.normal(-sd, sd, len(out["datetime"]))
#rnds = np.random.uniform(-2*np.pi, 2*np.pi, len(out["datetime"]))
#rnds = np.cos(rnds) * sd
out[v] = [m*j + a[v] + rnds[j-1] for j in i]
#out[v] = [m*j b[v] + rnds[j-1] for j in i]
#out["iaqAccuracy"] = 1
idf = pd.DataFrame(out)
reorder_columns = [col for col in out.keys() if col != 'datetime']
reorder_columns.append("datetime")
idf = idf.reindex(columns=reorder_columns)
#print(reorder_columns)
idf["year"] = [dt.year for dt in idf["datetime"]]
idf["month"] = [dt.month for dt in idf["datetime"]]
idf["day"] = [dt.day for dt in idf["datetime"]]
idf["hour"] = [dt.hour for dt in idf["datetime"]]
#idf["minute"] = [dt.minute for dt in idf["datetime"]]
idf["imputated"] = True
# original dataframe
idf = pd.concat([df.reset_index(drop=True), idf.reset_index(drop=True)])
idf.sort_values("datetime", inplace=True)
idf.reset_index(inplace=True, drop=True)
idf["datetime-1"] = idf["datetime"].shift(1)
idf["delta"] = idf["datetime"] - idf["datetime-1"]
idf["delta"] = idf["delta"].dt.seconds // 60**2
return idf
sinaica2 = sinaica_imputated.copy()
#sinaica2 = interpolate_missing(sinaica2, 1600)
#sinaica2[sinaica2.datetime >= pd.to_datetime('2021-03-15 09:00:00')][sinaica2.datetime <= pd.to_datetime('2021-03-17 11:00:00')]
imputation_list = [x for x in reversed(sinaica_imputated.delta[sinaica_imputated.delta > 1].index)]
sinaica2 = sinaica_imputated.copy()
for x in imputation_list:
try:
sinaica2 = interpolate_missing(sinaica2, x)
except:
print(f"Skipping {x}")
sinaica2 = sinaica2.loc[1:].reset_index(drop=True)
#sinaica2
Skipping 0 CPU times: user 759 ms, sys: 2.96 ms, total: 762 ms Wall time: 762 ms
airdata = pd.read_pickle("data/airdata/air-imputated.pickle.gz")
#airdata = airdata.groupby(["year", "month", "day", "hour"], as_index=False)
#airdata = airdata.aggregate(np.mean)
airdata = airdata[[c for c in airdata.columns if c not in ["iaqAccuracy", "year"]]]
#airdata["IAQ"] = airdata[["IAQ"]].round(0).astype('int64')
airdata["minute"] = airdata[["IAQ"]].round(0).astype('int64')
airdata = airdata[["datetime", "month", "day", "hour", "minute", "temperature", "pressure", "humidity",
"gasResistance", "IAQ"]].copy()
sinaica3 = sinaica2[[c for c in sinaica2.columns if c not in ["datetime-1", "delta",
"imputated", "year",
"datetime", "Fecha"]]]
sinaica3 = pd.merge(sinaica3, airdata, on=['month', 'day', 'hour'], how="left")
sinaica3 = sinaica3.loc[~sinaica3.isna().any(axis=1)]
sinaica3
| CO | NO | NO2 | NOx | O3 | PM10 | PM2.5 | SO2 | month | day | hour | datetime | minute | temperature | pressure | humidity | gasResistance | IAQ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 987 | 2.200000 | 0.205000 | 0.031000 | 0.207000 | 0.002000 | 45.000000 | 22.000000 | 0.004000 | 2 | 12 | 6 | 2021-02-12 06:05:35.846304417 | 35.0 | 21.51 | 777.41 | 44.04 | 152149.0 | 34.7 |
| 988 | 2.200000 | 0.205000 | 0.031000 | 0.207000 | 0.002000 | 45.000000 | 22.000000 | 0.004000 | 2 | 12 | 6 | 2021-02-12 06:05:38.837326527 | 34.0 | 21.51 | 777.41 | 43.98 | 152841.0 | 33.6 |
| 989 | 2.200000 | 0.205000 | 0.031000 | 0.207000 | 0.002000 | 45.000000 | 22.000000 | 0.004000 | 2 | 12 | 6 | 2021-02-12 06:05:47.812360048 | 32.0 | 21.54 | 777.41 | 43.73 | 153259.0 | 31.5 |
| 990 | 2.200000 | 0.205000 | 0.031000 | 0.207000 | 0.002000 | 45.000000 | 22.000000 | 0.004000 | 2 | 12 | 6 | 2021-02-12 06:05:50.803695202 | 32.0 | 21.53 | 777.41 | 43.70 | 152841.0 | 31.5 |
| 991 | 2.200000 | 0.205000 | 0.031000 | 0.207000 | 0.002000 | 45.000000 | 22.000000 | 0.004000 | 2 | 12 | 6 | 2021-02-12 06:05:53.795462847 | 30.0 | 21.52 | 777.41 | 43.70 | 153399.0 | 30.2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1582081 | 0.765217 | 0.009174 | 0.027826 | 0.039043 | 0.015304 | 20.304348 | 16.391304 | 0.001304 | 9 | 18 | 0 | 2021-09-18 00:59:47.142104626 | 138.0 | 26.00 | 782.92 | 56.34 | 916837.0 | 138.2 |
| 1582082 | 0.765217 | 0.009174 | 0.027826 | 0.039043 | 0.015304 | 20.304348 | 16.391304 | 0.001304 | 9 | 18 | 0 | 2021-09-18 00:59:50.136709690 | 138.0 | 26.00 | 782.92 | 56.33 | 917462.0 | 137.7 |
| 1582083 | 0.765217 | 0.009174 | 0.027826 | 0.039043 | 0.015304 | 20.304348 | 16.391304 | 0.001304 | 9 | 18 | 0 | 2021-09-18 00:59:53.131285429 | 138.0 | 26.00 | 782.90 | 56.34 | 916837.0 | 137.6 |
| 1582084 | 0.765217 | 0.009174 | 0.027826 | 0.039043 | 0.015304 | 20.304348 | 16.391304 | 0.001304 | 9 | 18 | 0 | 2021-09-18 00:59:56.125959396 | 136.0 | 26.00 | 782.92 | 56.35 | 921233.0 | 136.0 |
| 1582085 | 0.765217 | 0.009174 | 0.027826 | 0.039043 | 0.015304 | 20.304348 | 16.391304 | 0.001304 | 9 | 18 | 0 | 2021-09-18 00:59:59.120573282 | 134.0 | 25.99 | 782.92 | 56.35 | 922497.0 | 134.5 |
1581099 rows × 18 columns
sinaica3.to_pickle("data/sinaica/sinaica-imputated.pickle.gz")
We have imputated successfully all our data frame.
(sinaica2[sinaica2.delta > 2].
sort_values("delta", ascending=False)
[["Fecha", "delta"]].
reset_index(drop=True).
rename(columns={"delta": "Missing DAta", 'Fecha': 'Date'}).
head(10)
)
| Date | Missing DAta |
|---|
We recognize this might not be the best method, but we can explore more imputation methods on timeseries modeling.